Figure 2.
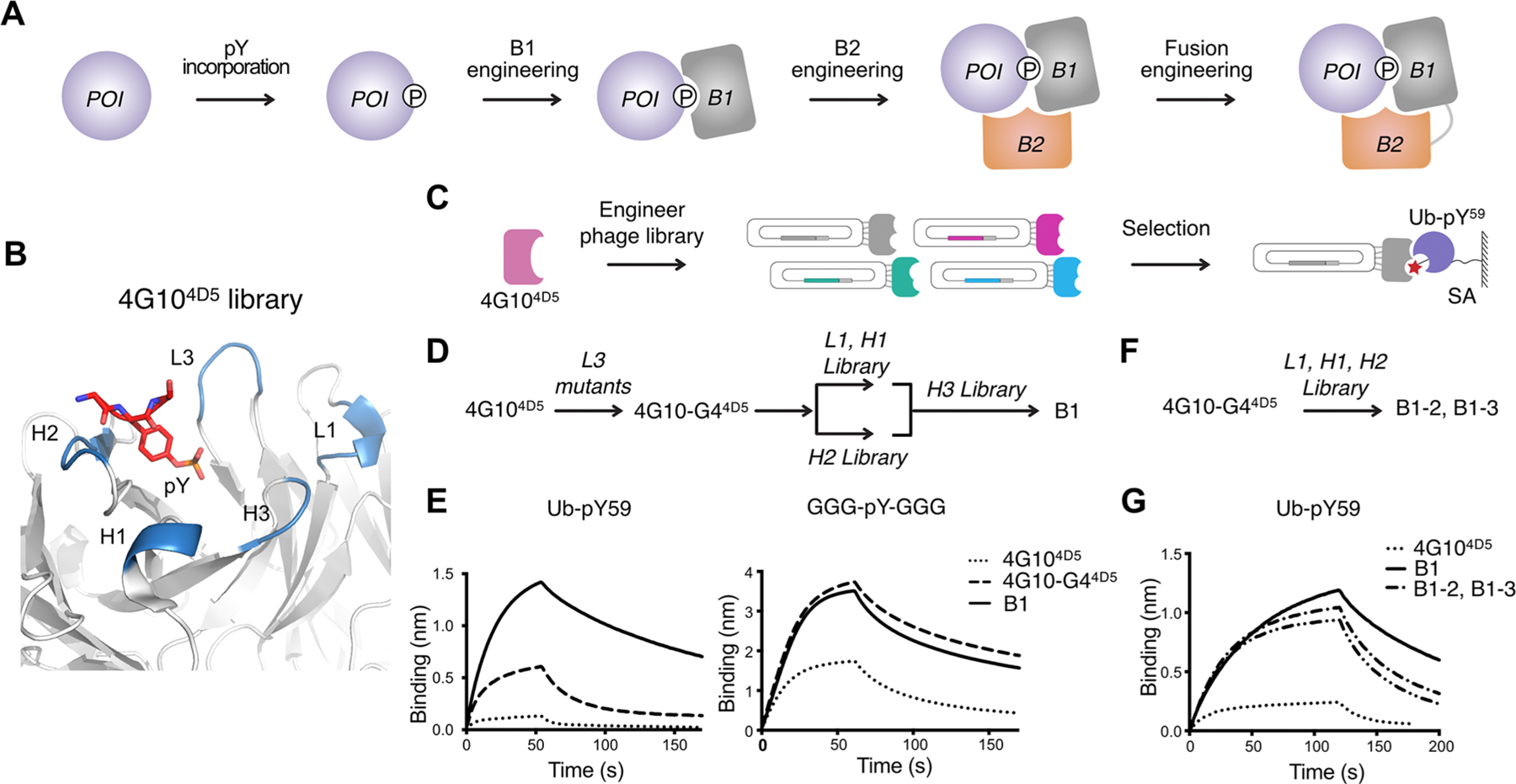
Anti-Ub-pY59 B1 engineering. (A) Four-step process to create pY-TRAPs to pY-native proteins: first, generate a protein of interest (POI) with the single pY modification; second, identify B1 that interacts with the pY motif and the surrounding sequence; third, identify B2 that conditionally binds to the complex of B1 and the POI at a composite site which includes both the POI and B1, and not either alone; and finally, engineer B1−B2 fusions to further improve binding affinity. (B) Structure of the 4G104D5 scaffold (PDB: 6DF1) showing the CDR loops (blue) and the pY residue (red). (C) Phage display workflow for anti-Ub-pY59 B1 selection. (D) CDR-walking approach to engineer anti-Ub-pY59 B1. (E) BLI characterization of 4G104D5, 4G10-G44D5, and B1 to Ub-pY59 and GGG-pY-GGG. (F) Single library approach where CDRs L1, H1, and H2 are simultaneously mutated to engineer anti-Ub-pY59 B1 binders. (G) BLI characterization of 4G104D5, B1, B1−2, and B1−3. The KDs of the interactions are summarized in Table S1.
