Figure 3.
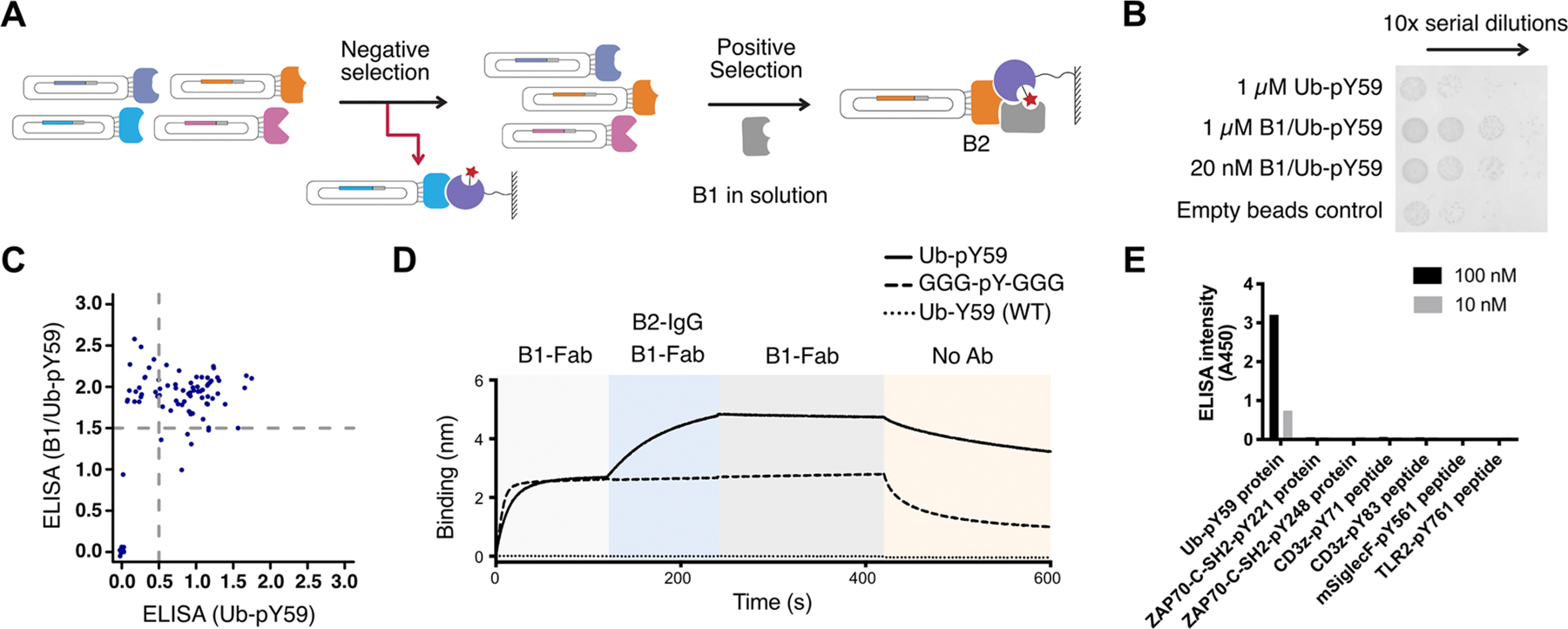
Anti-Ub-pY59 B2 engineering dramatically enhances affinity and specificity. (A) Phage display workflow for negative and positive selection for anti-Ub-pY59 B2. (B) Enrichment phage binders as a function of round by phage titer experiments. Binders that strongly interact with the B1/Ub-pY59 complex were more enriched than binders against Ub-pY59 as seen by higher numbers of phagemid colonies. (C) Characterization of binding of Fab-phage to Ub-pY59 or B1/Ub-pY59 in ELISA revealed clones that interact with both the Ub-pY59 and the B1/Ub-pY59 complex (upper right quadrant), and clones that selectively bind to the B1/Ub-pY59 complex (upper left quadrant). (D) Sequential BLI experiments show that B1 binds both Ub-pY59 (solid curve) and the GGG-pY-GGG control (dashed curve), while B2 added subsequently only recognizes the B1/Ub-pY59 complex (solid curve) but not the B1/GGG-pY-GGG complex (dashed curve); neither B1 nor B2 binds to the WT Ub-Y59 protein (dotted curve). (E) ELISA experiment showing that the anti-Ub-pY59 TRAP binders are highly selective toward Ub-pY59 and not other pY proteins or peptides such as the human ZAP70 SH-2 domain modified at two sites, CD3ζ modified at two sites, and TLR2 and murine SiglecF (mSiglecF) modified at one site.
