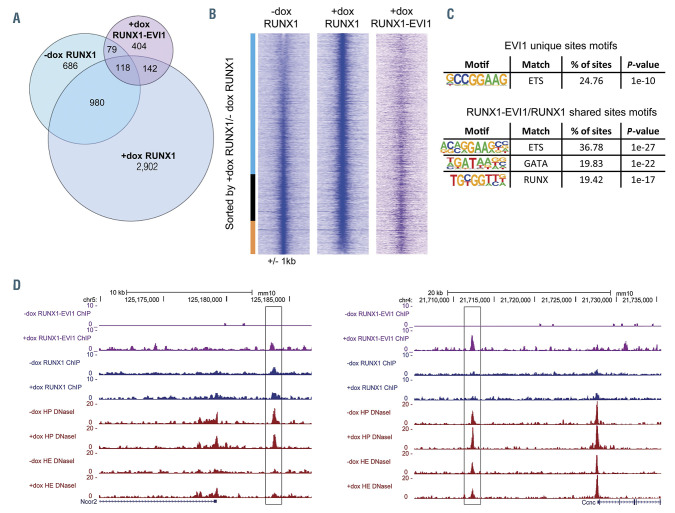
Figure 5.
RUNX1-EVI1 disrupts RUNX1 binding but also binds to unique binding sites. (A) Venn diagram showing the number of uniquely called and overlapping RUNX1 and RUNX1-EVI1 chromatin immunoprecipitation sequencing (ChIP-seq) peaks. (B) Comparison of RUNX1 binding in -dox and +dox treated cells, RUNX1 ChIPseq peaks were ranked according to the fold difference of the normalized plus doxycycline (+dox) /-dox tag count across a 2 kilobase (kb) window. The tag-density of the RUNX1-EVI1 ChIP-seq peaks is plotted alongside. The bar alongside indicates the +dox specific sites (blue), shared sites (black) and -dox specific sites (orange). (C) De novo motif enrichment was conducted within the RUNX1-EVI1 ChIP-seq peaks, at both the unique sites and those which were also bound by RUNX1 in either -dox, +dox or both. (D) Genome browser screenshots showing an example site where both RUNX1 and RUNX1-EVI1 bind (left, Ncor2 locus) and where RUNX1-EVI1 binds in the absence of RUNX1 (right, Ccnc locus).