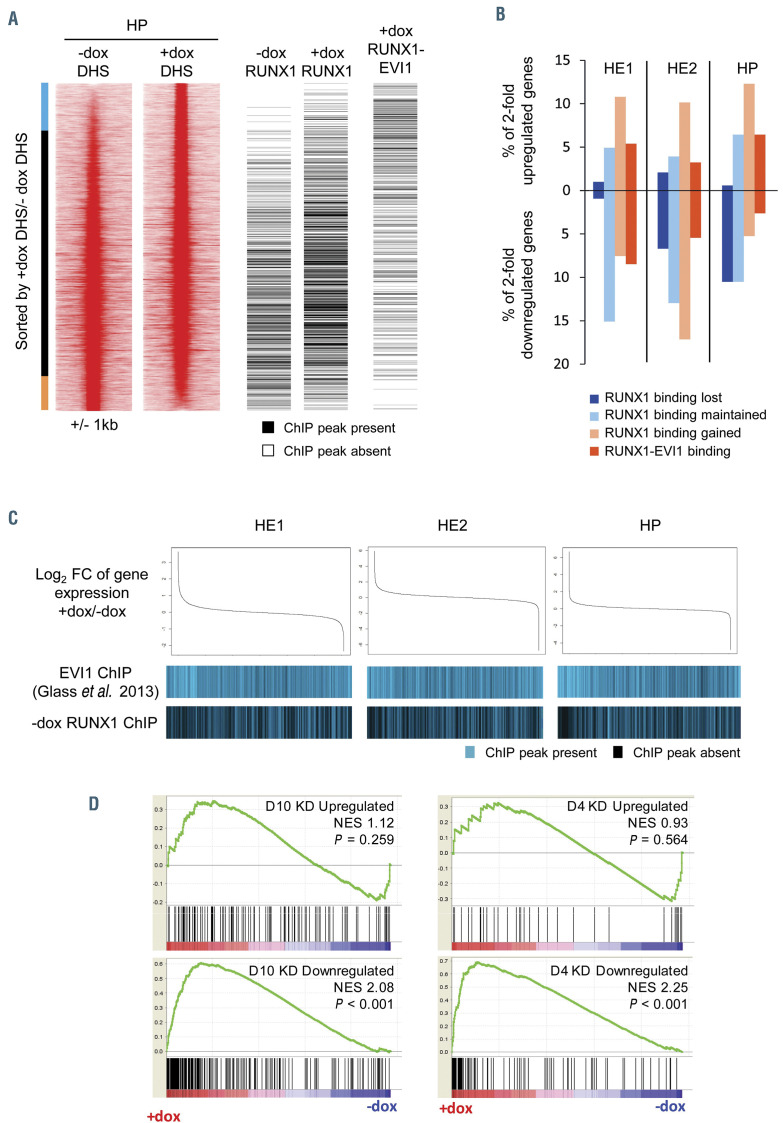
Figure 6.
Changes to chromatin organisation and gene expression are modulated by RUNX1 and RUNX1-EVI1 binding. (A) Comparison of RUNX1 and RUNX1-EVI1 binding sites to DNase I hypersensitive sites sequencing (DNaseI-seq) data from hematopoietic progenitor (HE) cells. DNaseI-seq peaks are ranked according to the fold-difference of the plus doxycycline (+dox)/-dox normalized tag-count, with the presence (black) or absence (white) of a chromatin immunoprecipitation sequencing (ChIP-seq) peak indicated alongside. The bar alongside indicates the +dox specific DNaseI sites (blue), shared sites (black) and –dox specific sites (orange). (B) The percentage of 2-fold de-regulated genes at each stage which have an associated RUNX1 or RUNX1-EVI1 binding site. (C) Comparison of changes in gene expression to the binding patterns of wild-type EVI1 and RUNX1. Gene expression was ranked by fold change (top), with the presence or absence of wild-type EVI127 or RUNX1 binding associated with each gene is indicated below in blue. (D) Gene set enrichment analysis comparing changes in gene expression after RUNX1-EVI1 induction in HP to genes that are up- and downregulated following 4 and 10 days of RUNX1-EVI1 knockdown in the SKH1 cell line.19 Genes that are upregulated following RUNX1-EVI1 induction correspond closely to genes that are downregulated after RUNX1-EVI1 knock-down in the SKH1 cell line.