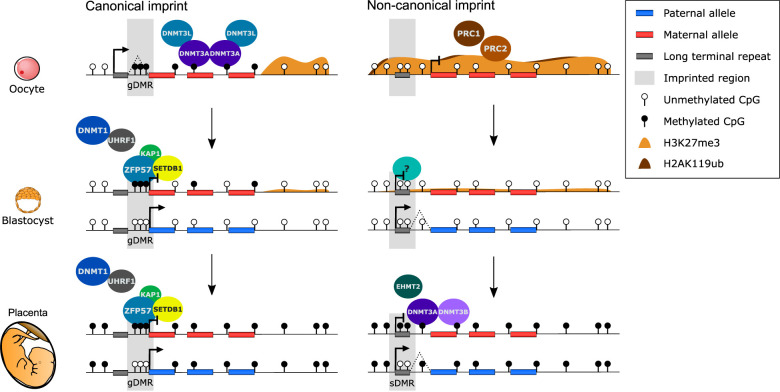
Figure 1.
Mechanisms of establishment and maintenance of maternal canonical and noncanonical imprinting. (Left) Canonical imprinting: DNA methylation is targeted to transcribed gene bodies, including canonical imprinted gDMRs, in oogenesis by tetramers of DNMT3A and DNMT3L. There is widespread usage of long terminal repeats (LTRs) as alternative upstream promoters in the oocyte. In the preimplantation and postimplantation embryo, a complex of ZFP57 (or ZNF445), TRIM28 (KAP1), and H3K9 methyltransferase SETDB1 localizes to gDMRs recruiting DNMT1 to maintain DNA methylation on the maternal allele. In the postimplantation embryo, the gDMR is present in the fetus and placenta, enabling imprinted gene expression of a single gene or cluster of genes. (Right) Noncanonical imprinting: H3K27me3 is established by PRC2, which is in part dependent on PRC1 ubiquitination of H2AK119, across untranscribed regions in oogenesis, including noncanonically imprinted LTRs. In the preimplantation embryo, maternal H3K27me3 is progressively lost genome-wide, and whether an unknown factor is required to mark the maternal allele of noncanonically imprinted LTRs remains unclear. In the postimplantation embryo, noncanonical imprints become placental specific, acquiring DNA methylation on the maternal allele, creating secondary DMRs (sDMRs), in placental and extraembryonic cell types. In the fetus (not shown), the noncanonically imprinted LTRs become biallelically methylated. The acquisition of DNA methylation in postimplantation development at these domains is dependent on EHMT2 activity, through either the deposition of H3K9me2 or post-translational modification of proteins integral for de novo DNMT activity.