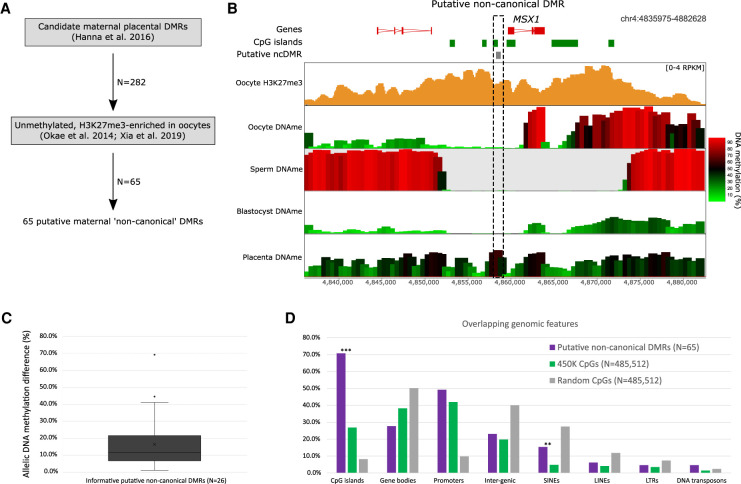
Figure 3.
Identification of putative noncanonical imprinted DMRs in the human placenta. Using the epigenetic patterning characteristic of noncanonical DMRs in mice, we investigated publicly available data to identify putative maternal “noncanonical” DMRs in humans. (A) Previously reported candidate maternal DMRs from placenta were evaluated for oocyte DNA methylation and H3K27me3, selecting those domains that were unmethylated and enriched for H3K27me3 in oocytes (N = 65). (B) Screenshot of putative noncanonically imprinted DMR upstream of the MSX1 gene. Enrichment for H3K27me3 in human oocytes is shown using running 500-bp windows, with a 100-bp step, quantitated as RPKM. DNA methylation in human oocytes, sperm, blastocyst, and first trimester placental trophoblast is shown using 1-kb running windows, with a 500-bp step. (C) The box plot shows the allelic difference in DNA methylation at informative putative noncanonical imprinted genes (N = 26). Informative DMRs were defined as those with at least three CpGs covered by at least two reads on each allele. (D) Overlapping genomic features were compared between putative noncanonical DMRs (N = 65) and CpGs on the Illumina 450K array (N = 485,512) using the χ2 statistic. P-value significance was adjusted for multiple comparisons using Bonferroni correction. (**) P < 0.001, (***) P < 0.0001. A random subset of genomic CpGs is shown in gray for context (N = 485,512).