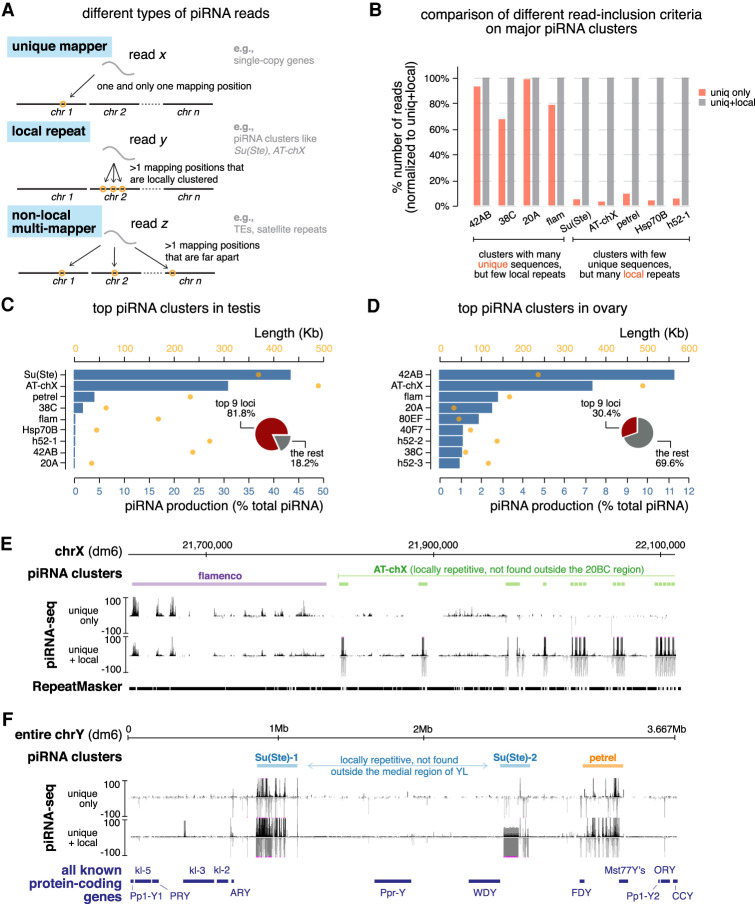
Figure 3.
Definition of piRNA clusters in testis and ovary using a new algorithm. (A) Three types of piRNA reads, defined based on their mapping positions. Uniquely mapped reads can be mapped to only one position in the genome and their origin is unambiguous. Reads derived from local repeats can be mapped to several positions in the genome; however, all of these mapping positions are locally clustered in a single genomic region. On the other hand, nonlocal multimappers can be mapped to multiple positions that are not restricted to one genomic region (typically mapped to more than one chromosome). Previously, only uniquely mapped reads were used to define piRNA clusters and quantify their expression, as the genomic origin of multimappers is ambiguous. Inclusion of multimappers derived from local repeats, as shown in this study, allows identification of new piRNA clusters as well as a more accurate quantification of piRNA production from known clusters. At the same time, it preserves the certainty that reads are generated from genomic loci in question. See Supplemental Figure S4 for detailed pipeline. (B) Histogram comparing numbers of mapped reads for major piRNA clusters using different read inclusion criteria as defined in A. For each cluster, the number of mapped reads generated by different methods is normalized to the method that includes both unique and local repeat reads (the right column). See also Supplemental Figure S4 and the Materials and Methods. (C) Expression of the top nine most active piRNA clusters in testis. Blue bars depict the contribution of each cluster to total piRNAs (percentage) and orange dots show cluster lengths according to the dm6 genome assembly. Insert is a pie chart of the contribution of the top nine loci to total piRNAs in testis. (D) Same as in C but for ovary. (E) UCSC genome browser view of a pericentromeric region (chrX) encompassing the entire flamenco locus (purple) and the distal part of AT-chX piRNA cluster (green). Below the genomic coordinates (dm6) are piRNA coverage tracks using different read inclusion criteria. Note that, whereas flamenco produces piRNAs that can be mostly mapped to unique genomic positions, AT-chX generates piRNAs that map to local repeats in this cluster, but nowhere else in the genome. (F) UCSC genome browser view of the entire Y chromosome that harbors two Su(Ste) loci (blue) and the novel petrel piRNA cluster (orange). piRNA coverage tracks using different read inclusion criteria are shown below genomic coordinates (dm6). At the bottom, all known Y-linked protein-coding genes are drawn for reference (not to exact scale). Note that piRNA profiles of Su(Ste) and petrel clusters collapse if piRNAs derived from local repeats are excluded.