Figure 5.
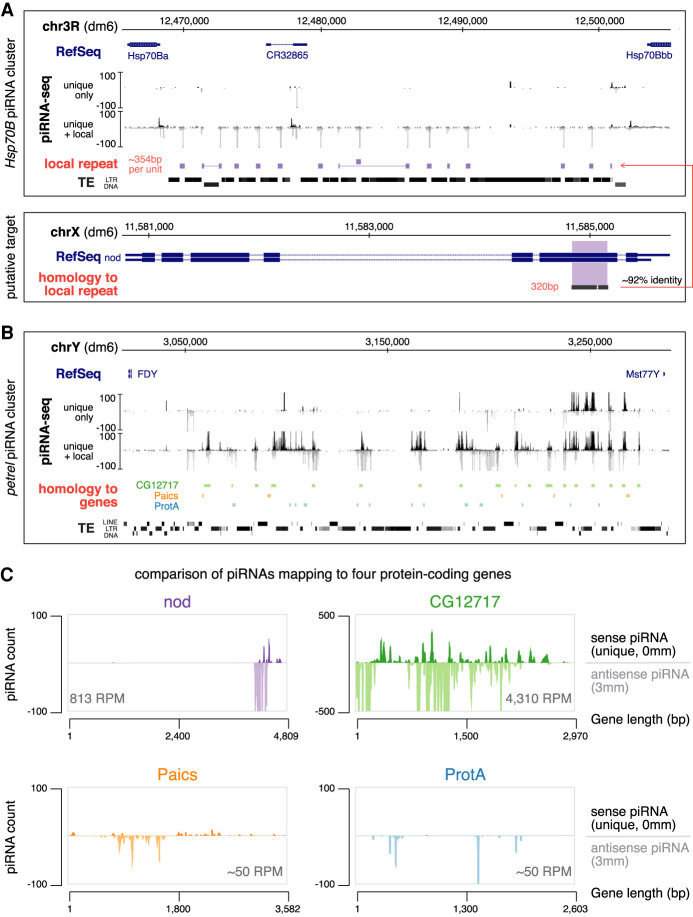
Hsp70B and petrel piRNA clusters encode piRNAs that target host genes. (A) Hsp70B piRNA cluster (top) and the putative target, nod (bottom). piRNA coverage tracks using different read inclusion criteria are shown below RefSeq and genomic coordinates (dm6) for the Hsp70B cluster. Approximately 354-bp local repeats homologous to a 320-bp exonic region of nod are depicted as solid blocks, which fill up most inter-TE space at this locus. Note that the “unique + local” piRNA track does not include TE-derived piRNAs that map outside this locus, but it picks up bona fide local repeats that are homologous, but not identical, to nod. (B) petrel piRNA cluster on the Y chromosome. piRNA coverage tracks using different read inclusion criteria are shown. Sequences with high levels of sequence similarity to protein-coding genes are depicted as colored blocks (not to exact scale). (Green) CG12717, (orange) Paics, (blue) ProtA. Note that gene homologous islands fill up most inter-TE space at this locus. Genomic coordinates are based on the dm6 genome assembly. (C) Coverage of sense (genome-unique, 0 mismatch) and antisense piRNAs (with up to three mismatches) over four putative, protein-coding gene targets of testis piRNAs. Antisense piRNA abundance is shown for each gene.