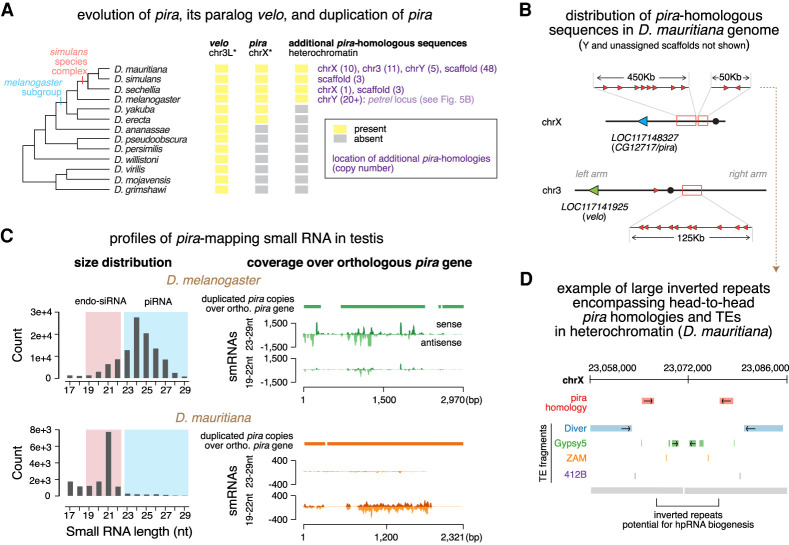
Figure 7.
Evolution of pira and pira targeting small RNAs. (A) Cladogram of major species in the Drosophila genus (left) and the evolutionary history of velo, pira, and pira-related sequences in genomes of these species (right). Orthologs were identified based on sequence homology and synteny. Shown in purple are locations of additional pira copies in each species and copy numbers in parentheses. Asterisk marks the chromosome name in the melanogaster subgroup, as karyotype differs in more distantly related species. (B) Cartoon depicting distribution of pira homologous sequences in the D. mauritiana genome. Orthologous pira is marked in blue, orthologous velo is marked in green, and the duplicated, candidate sources of pira targeting endo-siRNAs are marked in red. Note that they scatter across pericentromeric heterochromatin of chrX and chr3, as well as chrY and scaffolds (not shown). (C) Profiles of pira-mapping small RNAs in testes of D. melanogaster (top) and D. mauritiana (bottom). Size distributions are shown at the left. Coverage plots over the orthologous pira gene in each species are shown on the right, including: cumulative alignment of heterochromatic, duplicated copies of pira over the syntenic, orthologous pira (top; solid bar); stranded coverage of 23- to 29-nt piRNAs (middle; histogram); and 19- to 22-nt endo-siRNAs (bottom; histogram) over the orthologous pira gene. (D) Illustration showing two representative head-to-head copies of pira homology (red) in the pericentromeric heterochromatin of the D. mauritiana X chromosome. pira-related sequences are flanked by TEs and are part of a large inverted repeat that could potentially permit hpRNA biogenesis.