Figure 3.
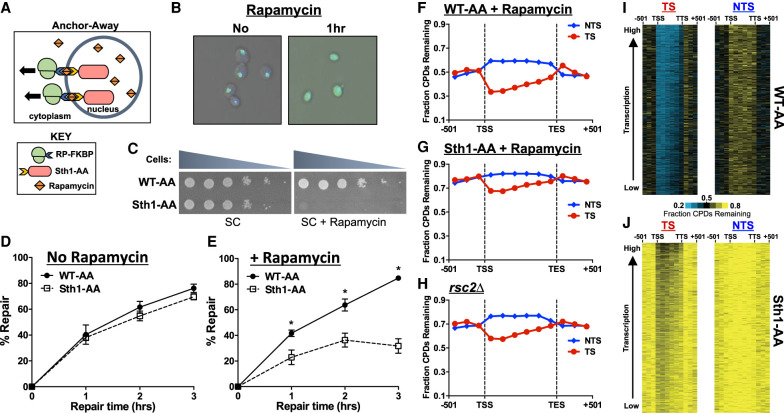
Depletion of RSC results in a genome-wide defect in NER. (A) Schematic of the anchor-away method. Ribosomal protein is tagged with a FKBP12 tag, whereas the nuclear protein of interest (Sth1) is tagged with a FRB tag. Upon treatment with rapamycin, the two tags rapidly dimerize, effectively depleting the nucleus of the protein of interest. (B) Microscopic confirmation of Sth1 depletion. The Sth1–anchor-away (Sth1-AA) protein is tagged with GFP, allowing for visualization of Sth1 trafficking from the nucleus to the cytoplasm following 1 h of rapamycin treatment. (C) Confirmation of Sth1 depletion via plating assay. Dilutions of WT-AA and Sth1-AA yeast were plated on synthetic complete (SC) and SC + rapamycin plates. The essential Sth1 subunit is successfully depleted on SC + rapamycin plates, resulting in cell death. (D,E) Quantification of T4 endonuclease V alkaline gel analysis of CPD repair for WT-AA and Sth1-AA with or without rapamycin treatment. The percentage of CPDs repaired at each time point is plotted as the mean ± SEM of a minimum of three replicates. P-values were calculated using an unpaired t-test with Holm–Sidak correction for multiple hypothesis testing. (*) P ≤ 0.05. (F–H) CPD-seq analysis examining the repair of CPDs on both DNA strands across approximately 5000 transcribed regions of the genome in WT-AA and Sth1-AA cells treated with rapamycin or in a rsc2Δ mutant. The fraction of CPDs remaining is calculated as the ratio of damage after 2 h of repair compared with the damage immediately following UV irradiation (0 h). (I,J) Gene plot analysis of WT-AA and Sth1-AA cells to examine CPD repair within individual genes on each DNA strand. The fraction of CPDs remaining following 2-h repair is plotted. Genes ordered based on transcription frequency (Holstege et al. 1998).