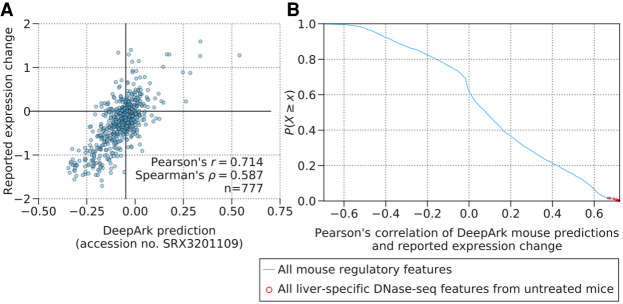
Figure 2.
DeepArk's variant effect predictions are well correlated with variant expression effects measured in a massively parallel in vivo reporter assay (MPRA) of enhancer activity. (A) The plot shows DeepArk's predictions for a liver-specific DNase-seq experiment (accession no. SRX3201109) for all possible variants (blue circles) in the ALDOB enhancer (hg19: Chr 9: 104,195,570–104,195,828) and the expression effects measured by the massively parallel reporter assay from Patwardhan et al. (2012), which are significantly correlated (Pearson's r = 0.714, P = 3.58 × 10−122, n = 777 and Spearman's ρ = 0.587, P = 2.91 × 10−73, n = 777). Note that the high degree of correlation with this DeepArk feature and the reported expression change is representative of the high correlation witnessed for liver-specific DNase-seq predictions in general, as shown in the next panel in comparison to other features. (B) The blue line in the plot shows the empirical complementary cumulative distribution function of the Pearson's correlation between the reported expression change in the MPRA of the ALDOB enhancer performed by Patwardhan et al. (2012) and DeepArk's variant effect predictions for each regulatory feature for M. musculus. The red circles correspond to liver-specific DNase-seq experiments from mice under control conditions (accession numbers SRX188645, SRX681492, SRX681493, SRX681494, SRX681495, SRX681496, SRX681497, SRX681498, SRX681499, SRX191053, and SRX3201109). The correlations of these liver-specific DNase-seq features are especially strong (average Pearson's correlation of 0.7046, n = 11 features), which is appropriate because the MPRA in question also measures the effects on expression levels in murine livers.