Figure 3.
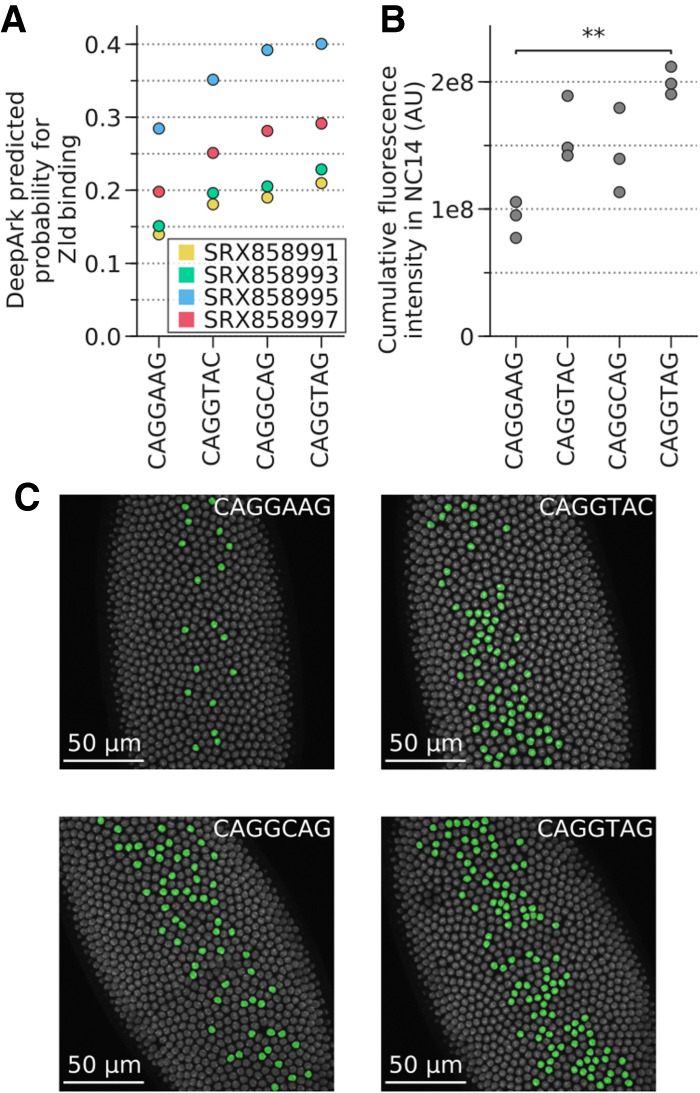
DeepArk's predicted effects for the different T48 mesodermal enhancer variants correlate with in vivo results. (A) Plot of all DeepArk predictions for Zld binding during nuclear cycle 14 for each of the four enhancer alleles. Each point represents a DeepArk prediction for a specific Zld ChIP-seq experiment and a specific allele. The CAGGTAG allele has the highest predicted probability of binding, whereas the reference allele CAGGAAG has the lowest. (B) The total transcriptional output for each of the four alleles, as quantified with in vivo MS2-GFP tagging during nuclear cycle 14. Each point in the graph represents the total transcription output (Methods) of all nuclei in a single embryo. Note that CAGGAAG and CAGGTAG have the lowest and highest transcriptional outputs, respectively, which is consistent with DeepArk's predictions. Bonferroni-corrected two-sided t-test with unequal variances: (**) P = 4.139 × 10−3; all others, P > 5 × 10−2. (C) False-color nuclei with active transcription in Drosophila embryos during minute 20 of nuclear cycle 14 illustrate the distinct levels of transcriptional activation induced by each allele (Supplemental Fig. S6). Three replicates were imaged for each allele. (AU) Arbitrary units, which represent the intensity of the pixels that correspond to the fluorescence of the MS2-GFP-tagged foci relative to the background GFP signal (Methods). (NC14) Nuclear cycle 14.
