Fig. 1. The SARS-CoV-2 pseudoknot interacts with the ribosome and pauses translation upstream of the slippery site.
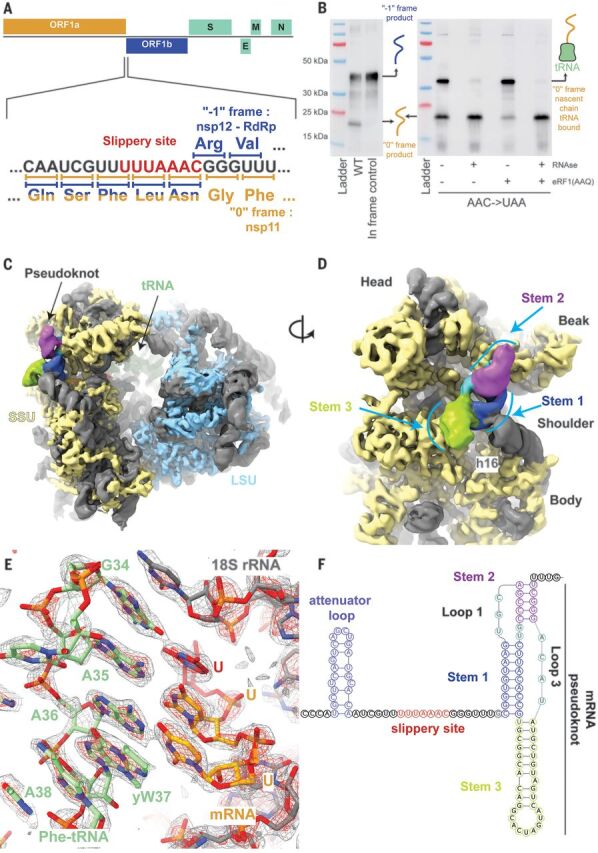
(A) Schematic of the SARS-CoV-2 main ORF. In the close-up view of the frameshift event, codons and corresponding amino acids are shown. During −1 frameshifting, the slippery site codons UUA (Leu) and AAC (Asn) are the last codons decoded in the 0 frame. Upon −1 frameshifting of the AAC codon to AAA, translation resumes at the CGG (Arg) triplet, where elongation proceeds uninterrupted to produce full-length Nsp12. (B) In vitro translation reaction depicting pausing at the frameshift site, as shown with Western blotting. Efficient frameshifting is observed for the WT template, consistent with our dual luciferase assays (see methods). Samples for cryo-EM originally intended to be trapped by dominant negative eRF1 (AAQ) show a tRNA-bound pause in proximity of the frameshift site. The tRNA-associated band is lost upon RNase treatment. Reactions without added eRF1 (AAQ) produce a similarly paused product. (C) Overview of the density low-pass filtered to 6 Å with the pseudoknot found close to the entry of the mRNA channel on the small subunit (SSU). The SSU proteins are colored in yellow, the large subunit (LSU) proteins in blue, and the rRNA in gray. The pseudoknot is colored according to its secondary structure as in (F), and the P-site tRNA is colored in green. (D) Close-up view of the pseudoknot from the solvent-exposed side of the SSU. Helix h16 of the 18S rRNA interacts with the base of Stem 1. Unpaired loop-forming nucleotides are colored in cyan. (E) P-site codon-anticodon interactions reveal a Phe (UUU) codon interacting with tRNA(Phe). yW37, wybutosine at position 37. (F) Schematic of the revised secondary structure elements in the pseudoknot necessary for −1 programmed ribosomal frameshifting, with different functional regions labeled and colored accordingly.
