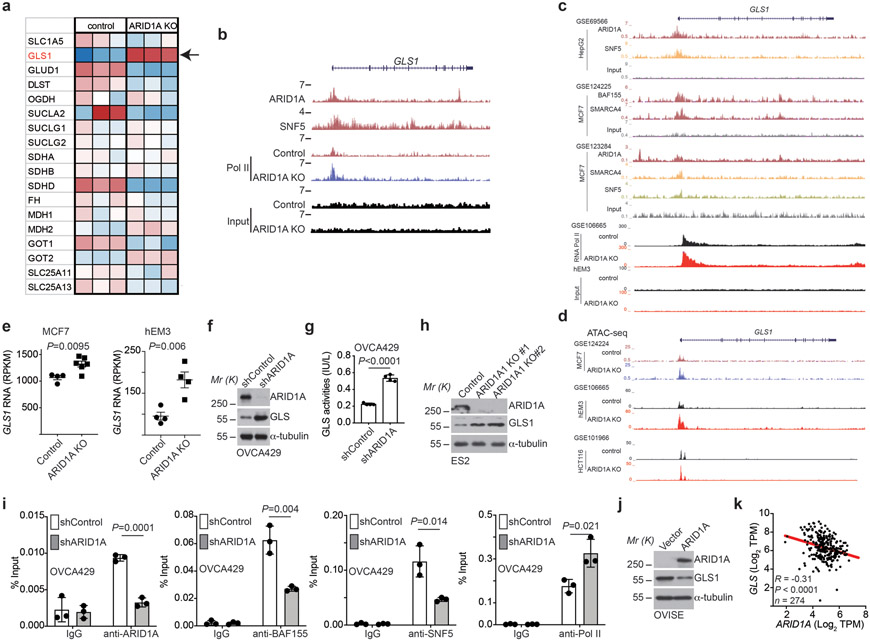
Extended Data Fig. 2. GLS1 is a direct target of the SWI/SNF complex.
a, Expression of glutamine metabolism related genes in control and ARID1A knockout RMG1 cells determined by RNA-seq analysis. Note that GLS1 shows the highest upregulation in response to ARID1A knockout. n=3 independent experiments. b, The indicated ChIP-seq and input tracks in the GLS1 gene locus in parental and ARID1A knockout RMG1 cells in previously published datasets (GSE120060). c, The indicated ChIP-seq and input tracks in the GLS1 gene locus in the indicated cancer cells based on the public database mining (GSE69566, GSE124225, GSE123284 and GSE106665). d, ATAC-seq tracks in the GLS1 gene locus in parental and ARID1A knockout cells based on the indicated datasets (GSE124224, GSE106665 and GSE101966). e, Expression of GLS1 mRNA in the indicated cancer cells based on based on mining public databases (GSE124227 and GSE106665). f-g, Control and ARID1A knockdown OVCA429 cells were examined for expression of ARID1A and GLS1 by immunoblot (f) or measured for glutaminase activity (g). n= 4 independent experiments. h, Control and ARID1A knockout ES2 cells were examined for expression of ARID1A and GLS1 by immunoblot. i, The association of ARID1A, BAF155, SNF5 and RNA Pol II with the GLS1 gene promoter in parental and ARID1A knockdown OVCA429 cells was examined by ChIP-qPCR analysis. An isotype matched IgG was used as a control. n = 3 independent experiments. j, Control and wildtype ARID1A ectopically expressing OVISE cells were examined for expression of ARID1A and GLS1 by immunoblot. k, Inverse correlation between GLS1 and ARID1A expression in 274 TP53 wildtype cancer cell lines across cancer types in the Cancer Cell Line Encyclopedia RNAseq database. Immunoblots are representative of three independent experiments with similar results in f, h and j. Error bars represent mean with s.d. in e, g and i. P value was calculated using two-tailed Student t-test in e, g, i and Spearman correlation analysis in k.