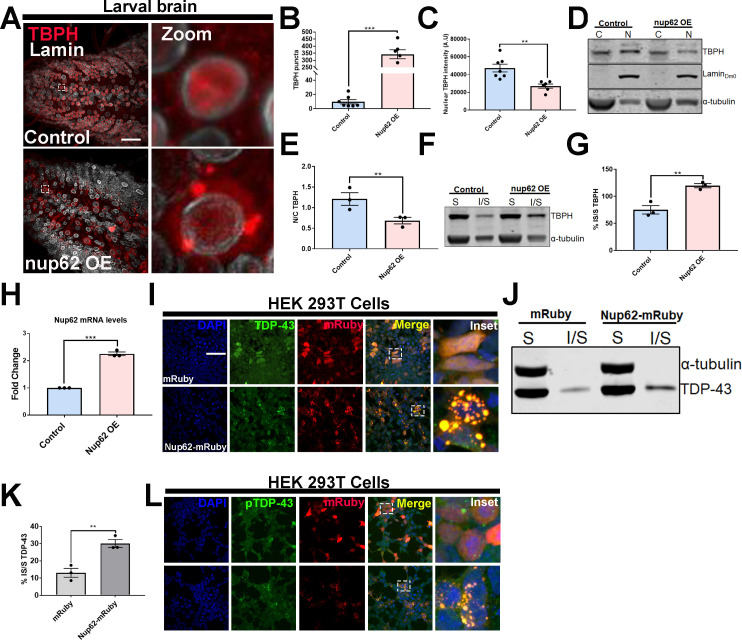
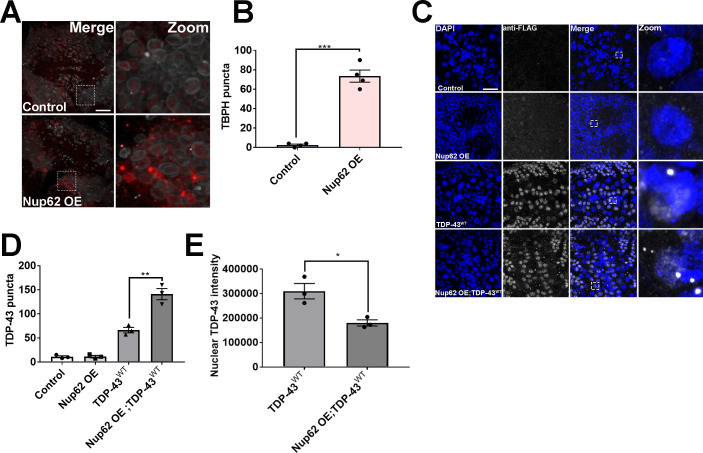
Figure 4. Nup62 expression promotes TDP-43 mislocalization and aggregation in vivo and in vitro.
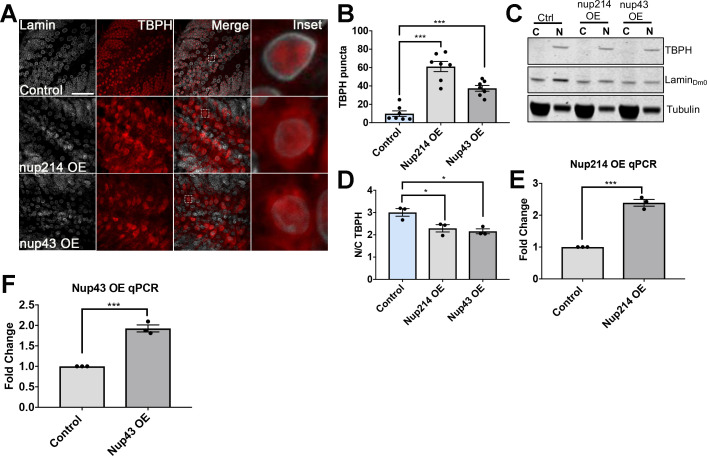
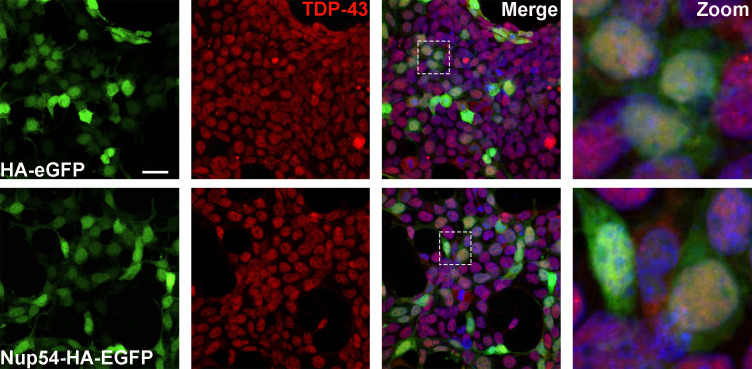
(A) Drosophila larval brain overexpressing Nup62 (Nup62 OE) in motor neurons (Ok371-gal4) and controls (eGFP) stained for the Drosophila homolog of TDP-43, TAR DNA-binding protein-43 homolog (Tbph), and the nuclear envelope marker Lamin, showing Tbph accumulation. Scale bar = 25 µM. Zoom image represents inset box. (B) Quantification of the number of Tbph puncta in Nup62 OE flies compared to controls (n = 5–7, ***p< 0.001). (C) Nuclear Tbph intensity in Nup62 OE flies compared to controls (n = 5–7 brains, **p < 0.01). (D) Western blots of cytoplasmic (C) and nuclear (N) fractions from Drosophila expressing Nup62 OE in motor neurons and control probed for Tbph, lamin, and tubulin. (E) Nuclear-cytoplasmic (N/C) ratio quantification of Tbph in Nup62 OE and control animals (n = 3 blots, **p < 0.01). (F) Western blots of soluble (S) and insoluble (I/S) fractions Drosophila expressing Nup62 OE in motor neurons and controls probed for Tbph and tubulin. (G) Percentage of Tbph solubility in Nup62 OE and control motor neurons (n = 3 blots, **p < 0.01). (H) qRT-PCR analysis of Nup62 mRNA in Nup62 OE animals compared to controls (n = 3, ***p < 0.001). (I) Representative immunofluorescence images of human embryonic kidney 293T (HEK293T) cells transfected with mRuby alone (red) and NUP62-mRuby show endogenous TDP-43 (green) coaggregation with NUP62. NUP62-mRuby co-localizes with endogenous TDP-43 (merge, yellow). Scale bar = 25 µM. (J) Western blots of soluble and insoluble fractions from HEK293T cells transfected with mRuby alone or NUP62-mRuby probed for TDP-43 and tubulin. (K) Quantification of TDP-43 solubility in HEK293T cells transfected with mRuby alone or NUP62-mRuby (n = 3 blots, **p < 0.01). (L) Representative immunofluorescence images of HEK293T cells transfected with NUP62 show phosphorylated TDP-43 accumulation (green) that co-localizes with NUP62 (merged, yellow). Scale bar = 25 µM. qRT-PCR and western blot analysis were done in triplicate using biological replicates. One-tailed t-test was used in panel B, C, E, G, H, and K. All quantification represent mean ± s.e.m.