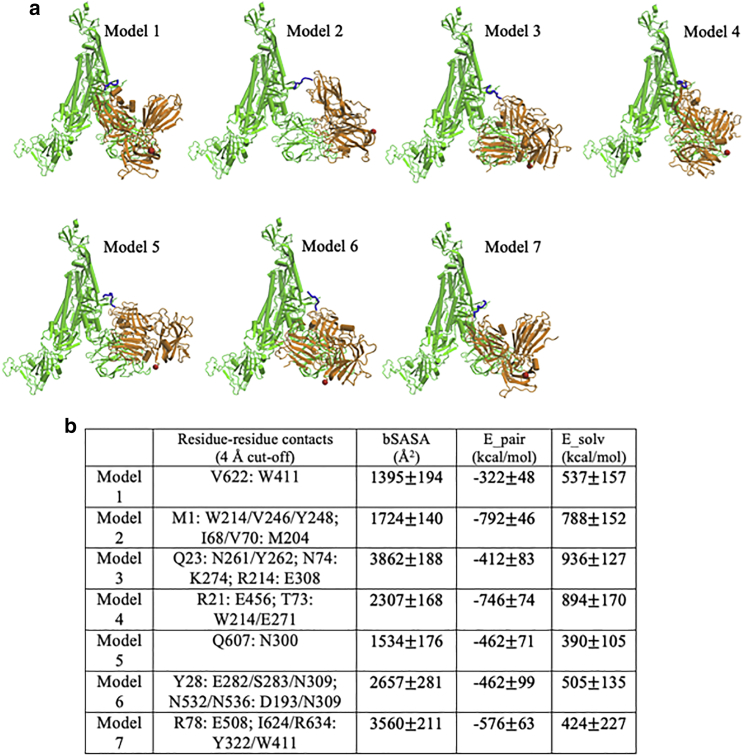
Figure 2.
Binding modes for seven predicted models. (a) Models, shown for Spike trimer (chain-C): Nrp1 (a2-b1-b2) binding. The TNSPRRAR motif is marked in blue. The last residue of the Nrp1 a2-b1-b2 domain segment is marked in red. (b) Noticeable residue-residue contacts (<0.4 nm) between Spike protein and Nrp1 in different models (except for the RRAR binding region). Together with additional residue-residue contacts (<0.6 nm), the proteins are seen to interact via domains a2, b1, and b2 on Nrp1 and a limited number of regions N1–N4, RBDN, RBDC, C1–C3, the very C-terminus of S1 and beginning of S2 (see Fig. S3 for details). Listed in Table b are the buried solvent-accessible surface area (SASA), pair-interaction energy, and changes of solvation energy for the different models. The interaction energies (based upon pairwise vdW and electrostatic forces) are calculated between Nrp1 (domains a2-b1-b2) with a full Spike trimer (chains A, B, and C). The models shown are listed in order of their population near their cluster centers (most to least). Standard errors were calculated over different frames of the simulation trajectories (saved at every 100 ps). To see this figure in color, go online.