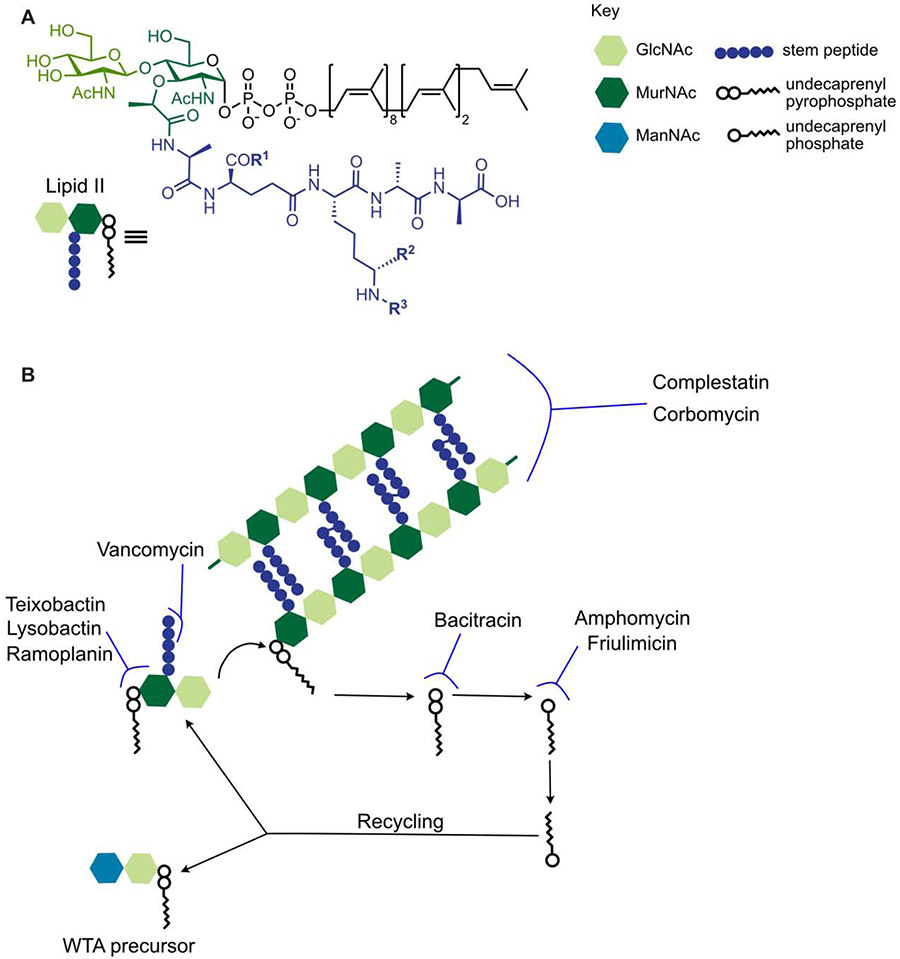
Figure 2: Substrate-binding antibiotics recognize various features of Lipid II, inhibiting different stages of peptidoglycan synthesis and lipid recycling.
A) The chemical structure of Lipid II. R1, R2, and R3 vary by species. In S. aureus, for example, R1 = NH2, R2 = H, and R3 = Gly5. B) When a Lipid II monomer is added to the existing PG matrix, undecaprenyl pyrophosphate is released. It is metabolized to undecaprenyl phosphate, which then gets flipped across the membrane for recycling. This lipid carrier is used in both the PG monomer, Lipid II, and in WTA precursors. Teixobactin, ramoplanin, and lysobactin bind the hydrophilic head group of Lipid II, vancomycin binds the stem peptide, and corbomycin and complestatin are proposed to bind formed PG. Bacitracin inhibits the metabolism of undecaprenyl pyrophosphate to undecaprenyl phosphate, preventing recycling of this lipid carrier, while amphomycin and friulimicin bind undecaprenyl phosphate.