Figure 4. The GH0XJ048933 SE codes for an eRNA and co-regulates the genes of the GRIPAP1/PIM2 locus.
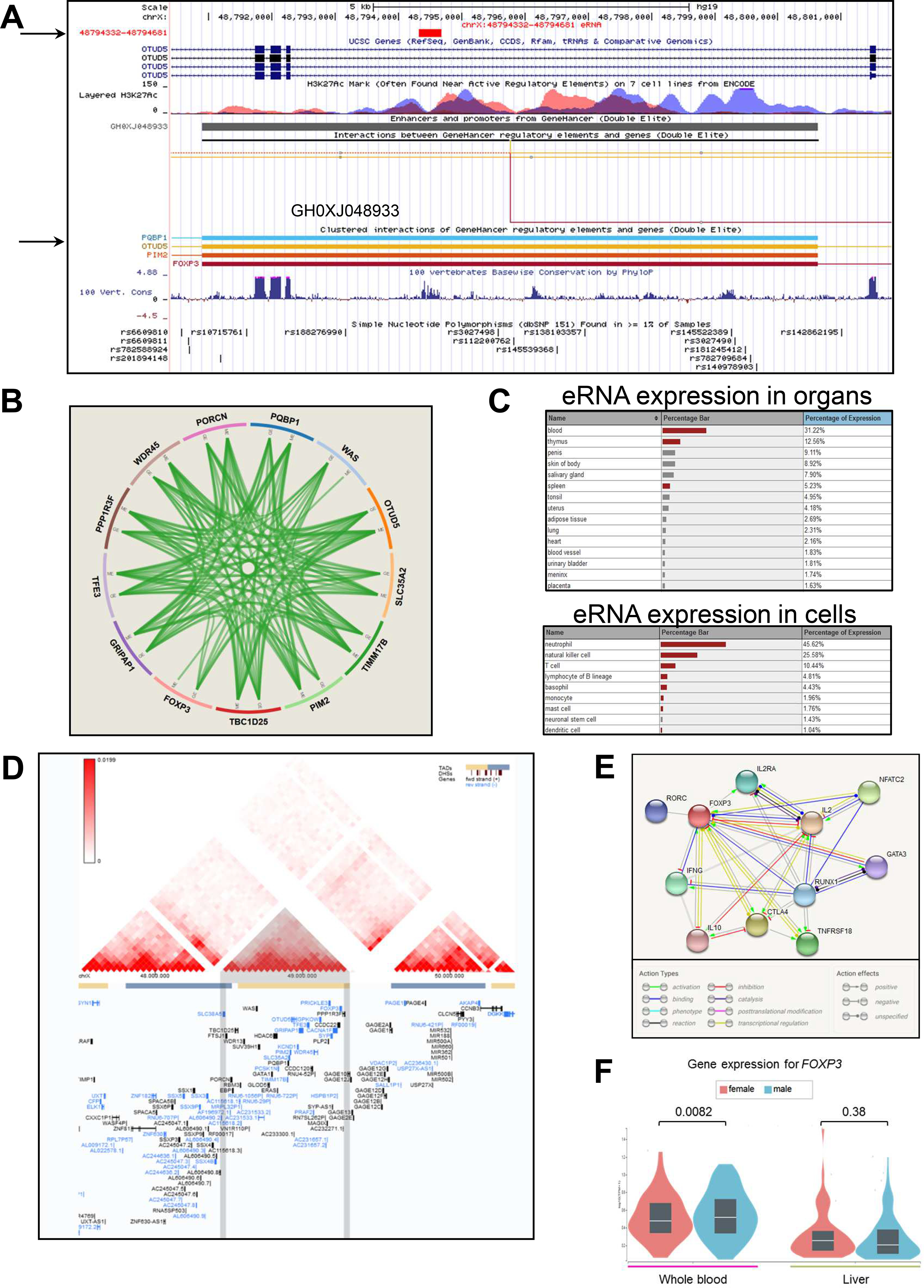
A) Screenshot from the UCSC Genome browser showing the GH0XJ048933 SE region (chrX:48,791,000–48,802,000). Listed tracks are: i) the ruler with the scale at the genomic level; ii) chrX nucleotide numbering; iii) the track for eRNAs from the FANTOM5 Human Enhancers project (http://slidebase.binf.ku.dk/human_enhancers/); iv) the UCSC RefSeq track; v) ENCODE data for H3K4Me1, H3K4Me3, H3K27Ac histone marks, derived from seven cell lines; vi) the enhancers/promoters track from GeneHancer;41 the grey bar indicates the GH0XJ048933 enhancer; vii) interactions connecting GeneHancer regulatory elements and genes (interactions with OTUD5, PIM2, PQBP1, FOXP3 are depicted); viii) basewise conservation track; ix) the dbSNP(151) track for common polymorphisms.
B) Integration of gene expression (GE), protein expression (PE), copy number (CN) and methylation (ME) relative to the 13 genes regulated by the GH0XJ048933 SE. Data come from the TCGA portal (https://tcga-data.nci.nih.gov/docs/publications/tcga/?). Circle plot was built by using the Zodiac tool (http://www.compgenome.org/zodiac/).42 Only significant intergenic interactions are shown (FDR≤0.1). Green lines indicate positive interactions.
C) The tables show expression data (>1%) in organs/cells for the eRNA gene mapping within GH0XJ048933. Red bars indicate a significant over-representation of the transcript (FANTOM5 data).
D) TAD structure of the chrX:47,480,000–50,440,000 region. The central TAD contains all genes of the PBC-associated region tagged by rs7059064. The panel was produced though the 3D-Genome Browser (http://3dgenome.org),43 using Hi-C data produced in HepG2 cells (hepatocytes) and generated by the Dekker Laboratory (resolution: 40kb).
E) FOXP3 interactome. The best 10 interactions are shown (highest confidence=90%). Evidence are based on text-mining, experiments, databases, co-expression data, gene fusions, co-occurrences. The panel was produced using the STRING tool (https://string-db.org/).
F) Violin plots show FOXP3 RNA expression levels in whole blood and liver, obtained through the GTEx portal, stratified on sex (265 males, 142 females).
