Abstract
The structure, size, and main physicochemical characteristics of the SARS-CoV-2 virion with the spike transmembrane protein corona were discussed. Using these data, diffusion coefficients of the virion in aqueous media and in air were calculated. The structure and dimensions of the spike protein derived from molecular dynamic modeling and thorough cryo-electron microscopy measurements were also analyzed. The charge distribution over the molecule was calculated and shown to be largely heterogeneous. Although the stalk part is negatively charged, the top part of the spike molecule, especially the receptor binding domain, remains positively charged for a broad range of pH. It is underlined that such a charge distribution promotes the spike corona stability and enhances the virion attachment to receptors and surfaces, mostly negatively charged. The review is completed by the analysis of experimental data pertinent to the spike protein adsorption at abiotic surfaces comprising nanoparticle carrier particles. It is argued that these theoretical and experimental data can be used for developing quantitative models of virus attachment to surfaces, facilitating adequate analysis of future experimental results.
Keywords: Adsorption of spike protein, Charge distribution over spike protein, Receptor attachment of spike protein, SARS-CoV-2 virion, Spike protein, Structure of SARS-CoV-2 virion
Introduction
Within a short time after the outbreak of the COVID-19 pandemic, an avalanche of publications have appeared, which are devoted to various aspects ranging from the SARS-CoV-2 virus transmission mechanisms [1, 2, 3, 4∗∗], its attachment to receptors and abiotic surfaces (fomites) [5∗∗, 6∗, 7, 8], inactivation and removal [8,9] to vaccination [10].
For the sake of conciseness, in this fragmentary review, attention is focused on the physicochemical characteristics of the virus particle (virion), especially the spike protein corona, which determines its attachment efficiency to various surfaces. It is hypothesized that viruses can be treated as composite nanoparticles characterized by well-defined physicochemical properties. Therefore, it can be assumed that their transfer to various surfaces comprising airborne particulate matter surfaces is a phenomenon analogous to colloid particle deposition processes. This concept was quantitatively elaborated in a recent review [4], where it is shown that the virus transmission via particulate matter or aerosol droplets acting as a shuttle can play a significant role. Therefore, in analogy to the colloid deposition, one can distinguish three main steps of virus particle (referred to as virion) attachment [4]: (i) transfer over macroscopic distances to the vicinity of boundary surfaces, either abiotic, for example, masks, or cell membranes, which is governed by forced convection (flow) and diffusion, for example, through mucus layer, (ii) transfer through the thin surface layer adjacent to interfaces which is controlled by the electrostatic and van der Waals forces (the latter are often referred to as hydrophobic interactions), and (iii) formation of a physical contact of the virion with the interface or the corresponding cellular receptor, where except for the mentioned interactions, the hydrogen bonding may play a significant role. One should mention that in the latter case, there appears another important step consisting in the penetration of the virion through the membrane into the host cell.
The overall virion attachment rate, quantified in terms of the solute flux, is controlled by the slowest step, often the transfer in the bulk phase, that is, by the virus concentration in the ambient phases. However, the actual (maximum) number of attached virions is governed by the surface binding energy, that is, affinity to receptors. The latter is controlled by the external layer of the virion, consisting of, the corona of spike proteins bearing appropriate binding sites. Considering this, one can argue that a quantitative analysis of the attachment kinetics requires thorough information about physicochemical parameters characterizing the virion and the spike protein. Therefore, in the first part of the review, the SARS-CoV-2 virion architecture with emphasis on the spike protein structure and its physicochemical characteristics including the heterogeneity of charge distribution is analyzed. Then, recent results derived from molecular dynamic modeling and experimental investigations pertinent to the spike protein attachment to receptors and adsorption at biotic surfaces are discussed. It is argued that these theoretical and experimental data discussed in this work can be used for developing quantitative models of virus attachment kinetics to receptors and abiotic surfaces, facilitating adequate analysis of future experimental data.
Virion structure and physical parameters
SARS-CoV-2 is an enveloped virus belonging to the coronaviridae family that causes respiratory and enteric system infections in humans and many animals. It shows similarity to SARS-CoV, a coronavirus known since 2003 [11] in respect to the structure of the membrane and the spike protein shell [5,6,12,13]. In addition, it uses the same ACE2 receptor for the attachment to cells. This similarity allows drawing some conclusions about its structure and mechanism of action.
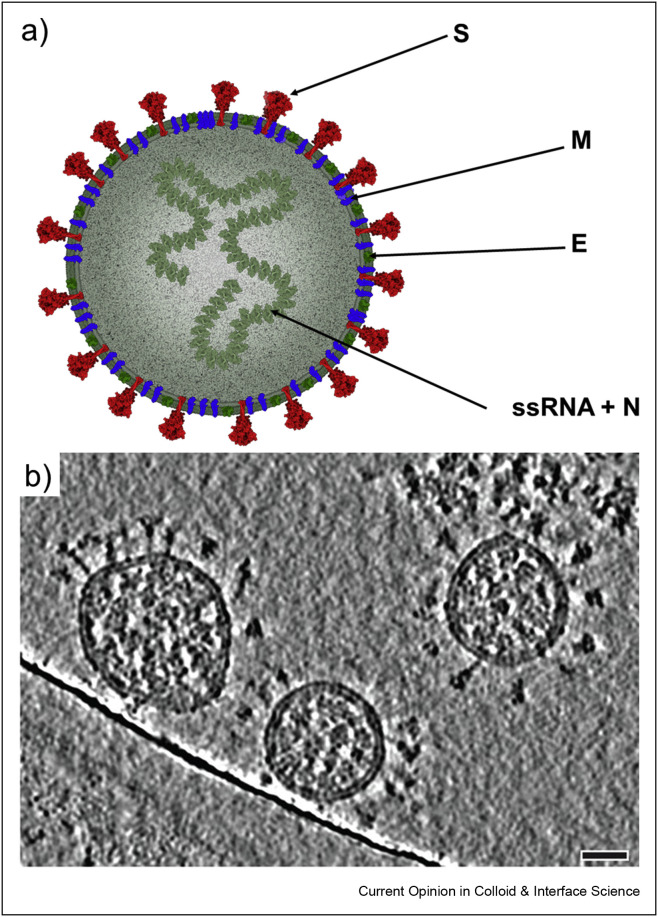
The SARS-CoV-2 virion schematically shown in Figure 1 contains a single-stranded RNA genome composed of ca. 30,000 base pairs. The genome is encapsidated by the N protein. On the other hand, the envelope of the virion comprises the M (membrane) protein that plays an important role in all coronavirus assemblies [11] and the E protein. The spike (S) protein incorporated into the membrane (see Figure 1) is the virus receptor protein also responsible for the virus and the cell membrane fusion. Molar masses of proteins are as follows: E—8.4, M—25.1, N—45.6, and S—141.2 kg mol−1 [11,16]. The main structural and conformational information about the virion stems from thorough cryo-TM imaging [15,17,18]. On the other hand, the virion micromechanical properties comprising its compliance and resistance to temperature perturbation were determined by atomic force microscopy (AFM) imaging and the force spectroscopy measurements [19]. These experiments enabled to establish that the virion is spherical in shape (see Figure 1) characterized by the core external dimeter equal to 91 ± 11 nm (an average value taken from 179 single particles [18]) with the membrane thickness of 5 ± 1 nm [15,20]. The AFM measurements are less accurate because of tip convolution effects yielding the adsorbed virion height equal to 60 nm [19]. Assuming a spherical shape of the virion, the core part surface area is equal to 2.6 × 104 nm2 (see Table 1 ). One can also calculate the virion diffusion coefficients in various media using the Stokes–Einstein relationship,
| (1) |
where k is the Boltzmann constant, T is the absolute temperature, η is the medium viscosity, and d H is the hydrodynamic diameter.
Figure 1.
A schematic representation of a SARS-CoV-2 particle (a), obtained using CellPAINT software [14], slices through tomographic reconstructions of SARS-CoV-2 virions (b), reprinted with permission from Turoňová et al. [15], scale bar, 30 nm.
Table 1.
Primary physicochemical characteristics of SARS-CoV-2 virion and its spike protein.
| Quantity (unit), symbol | Value | Remarks |
|---|---|---|
| Virion core part diameter (nm), dc | 91 ± 11 | Ke et al. [18], cryo-EM |
| Membrane thickness (nm), dm | 5 ± 1 | Turoňová et al. [15], cryo-EM |
| Core surface area (nm2), Sc | 2.6 × 104 | Calculated as |
| Virion diffusion coefficient, D (m2 s−1) | 2.7 × 10−10 5.4 × 10−12 | Air (298 K) Aqueous media (298 K) Calculated from the Stokes–Einstein relationship |
| S-protein Molar mass (kg mol−1), |
141.2 kg mol−1 | Ref. [16] |
| Density (kg m−3) | 1.35 × 103 | Ref. [23] |
| Molar volume, (nm3) | 1.7 × 103 | Calculated as |
| Equivalent sphere diameter, ds (nm) | 15 | Calculated as |
| Diffusion coefficient Ds (m2 s−1) | 3.3 × 10−11 | Aqueous media (298 K) Calculated from the Stokes–Einstein relationship for the equivalent sphere |
| Length (nm), dl | 24 ± 9 ~28 (~26) |
Ke et al. [18], cryo-EM Amaro et al. [22], ‘open’ (‘closed’) state |
| Number, per virion, N (1) | 26 40 61 |
Ke et al. [18], cryo-EM Turoňová et al. [15], cryo-EM Kiss et al. [19], AFM |
| Surface concentration, Ns (nm2) | 1–2.3 × 10−3 | Calculated as |
| Cross-sectional area at the top, Ss (nm−2) | 97 110 |
Closed conformation Open conformation This work (Figure 2(b)) |
| Coverage at the core, θs (dimensionless) |
0.097–0.22 0.11–0.25 |
Closed conformation Open conformation Calculated as NsSs |
Assuming that the hydrodynamic diameter of the virion corresponds to its core part diameter one obtains from Eq. (1), D = 2.7 × 10−10 and 5.4 × 10−12 m2 s−1 for the air and aqueous phases, respectively (at the temperature of 298 K). Obviously, in other media characterized by a larger viscosity, such as, for example, saliva or the mucus [21], the virion diffusion coefficient is significantly smaller, creating additional resistance to the overall virion transfer rate.
As mentioned, the virion membrane comprises a number of spike proteins, which exhibit a large conformational flexibility as demonstrated experimentally [15] and by extensive molecular dynamics modeling [22]. The distribution of the spike protein molecules over the core and their number were quantitatively evaluated by cryo-TM [15,18] and by AFM [19]. It is confirmed that the number of molecules per virion, denoted by N , is characterized by a large spread with the average values ranging between 26 [18] and 61 [19]. This effect may be caused by the disruption of the protein/membrane contact on freezing. It is interesting to mention that the spike protein molecule orientation is rather variable with the average angle of the longer axis toward the membrane equal to approximately 50 deg. [18] Using the N numbers, one can calculate that the spike protein surface density varies between 1 × 10−3 and 2.3 × 10−3 nm2. However, one should mention that at present, our knowledge about the membrane surface charge density as a function of pH and its ion adsorption properties is fairly limited.
Summarizing the abovementioned data, one can approximate the SARS-CoV-2 virion as a spherical core/shell nanoparticle with heterogeneous charge distribution as demonstrated in the study by Ghosh Chaudhuri and Paria, and Adamczyk et al. [24,25].
The structure and physicochemical parameters pertinent to the spike protein are analyzed in more detail in the next section.
The spike protein
Structure and dimensions
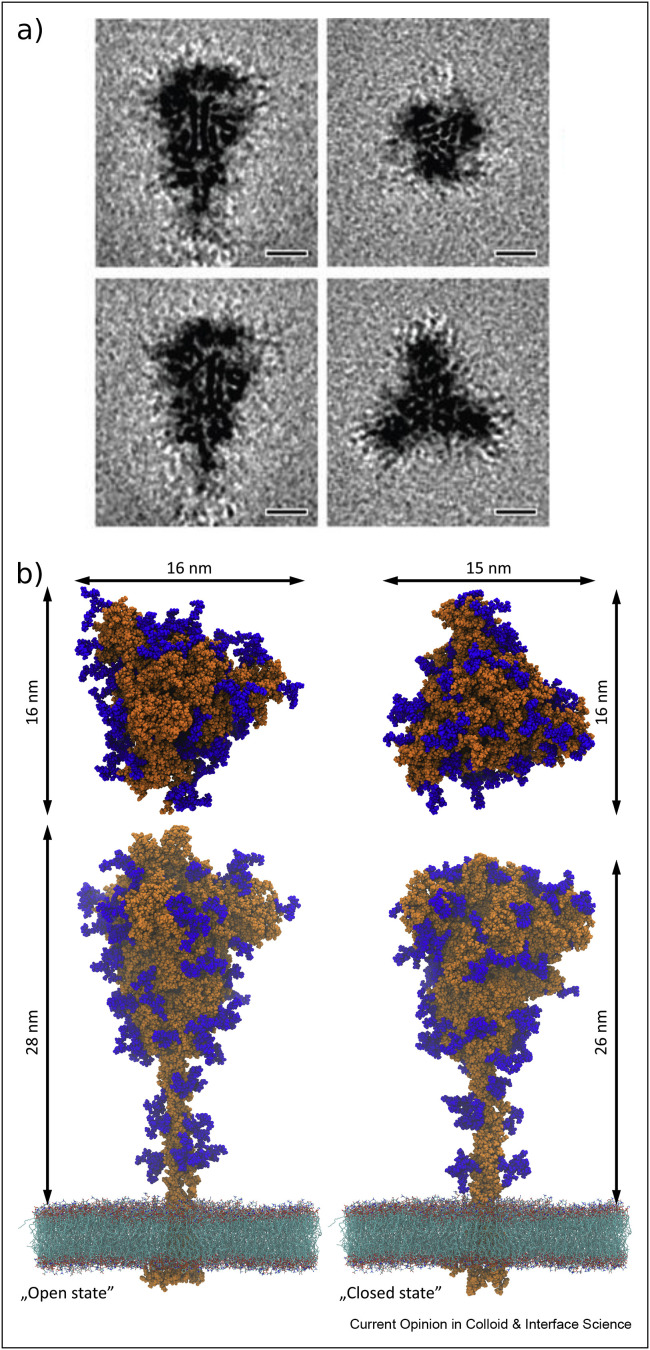
Detailed information on structure and function of the S protein is given in the study by Turoňová et al., Casalino et al., and Walls et al. [15,22,26]. In brief, it represents a viral fusion protein with a club-like shape of approximately 25 nm in length, as confirmed by cryo-EM measurements, see Figure 2(a) [15]. Precise determination of the S protein dimension can be obtained via molecular modeling performed using the protein crystal structure. Amaro et al. [22] have modeled SARS-CoV-2 spike glycoprotein in a realistic membrane via microsecond-long, all-atom molecular dynamics simulation. They assumed pH 7.4 (protonation states were estimated using PROPKA3 [27]) and 150 mM NaCl concentration to mimic the physiological conditions. It is shown that glycans cover a vast amount of the S protein surface area and relate this change with the open and closed conformational state of the protein. This highlights the shielding role of protein glycosylation in viral pathogenesis via forming thick N-glycan coating of the viral fusion proteins. In the case of the SARS-CoV-2 virion, the N-glycans are densely packed and effectively decrease protein accessible surface area, they account to ca. 20% of the S glycoprotein molar mass. Interestingly, Amaro et al. [22] also observed the reduced ACE2 binding when specific N-glycans were removed, suggesting their important role in the binding mechanism.
Figure 2.
Cryo-EM subtomogram average of the ectodomain of S protein shown as slices through the reconstruction (a), reprinted with permission from Turoňová et al. [15], scale bars, 5 nm. The original visual representation of S glycoprotein derived based on molecular structure provided in the study by Casalino et al. [22] with marked dimensions (b). Atoms are presented as spheres of Van der Waals radius. Protein and N-glycans are highlighted in orange and blue, respectively.
The molecular structure obtained in the study by Casalino et al. [22] for open and closed conformation is used to visualize and to determine the dimensions of the SARS-CoV-2 spike protein, shown in Figure 2 (b). This enables estimation of the most relevant physiochemical parameters, such as the protein number surface density, cross-sectional area, or its surface coverage, see Table 1. These primary physicochemical data can also be used for calculating several derivative parameters characterizing the protein, which facilitate a proper interpretation of experimental results. Thus, considering that the molar mass (denoted by M s) is equal to 141 kg mol−1 and the protein density ρ s is equal to 1.35 × 103 kg mol−1 [16,23], one can directly calculate that its specific molecule volume (where Av is the Avogadro constant) is equal to 1.7 × 103 nm3 (Table 1). Using this value, one can calculate the diffusion coefficient of the free spike protein molecule in aqueous media from Eq. (1). The hydrodynamic diameter can be calculated approximating the molecule by a prolate spheroid from the following formula
| (2) |
where a and b are the longer and the shorter spheroid semiaxes connected with the spheroid volume (equal to the molecule volume) through the dependence
| (3) |
where = a/b is the aspect ratio of the equivalent spheroid.
From Eqs. (2), (3), one obtains for a sphere and 15.4 nm for a spheroid characterized by the aspect ratio of two. Interestingly, this agrees with the dimension of the spike protein molecule at the top (see Figure 3 ). One can also deduce from this estimation that the elongated shape of the spike protein molecule little affects its hydrodynamic diameter.
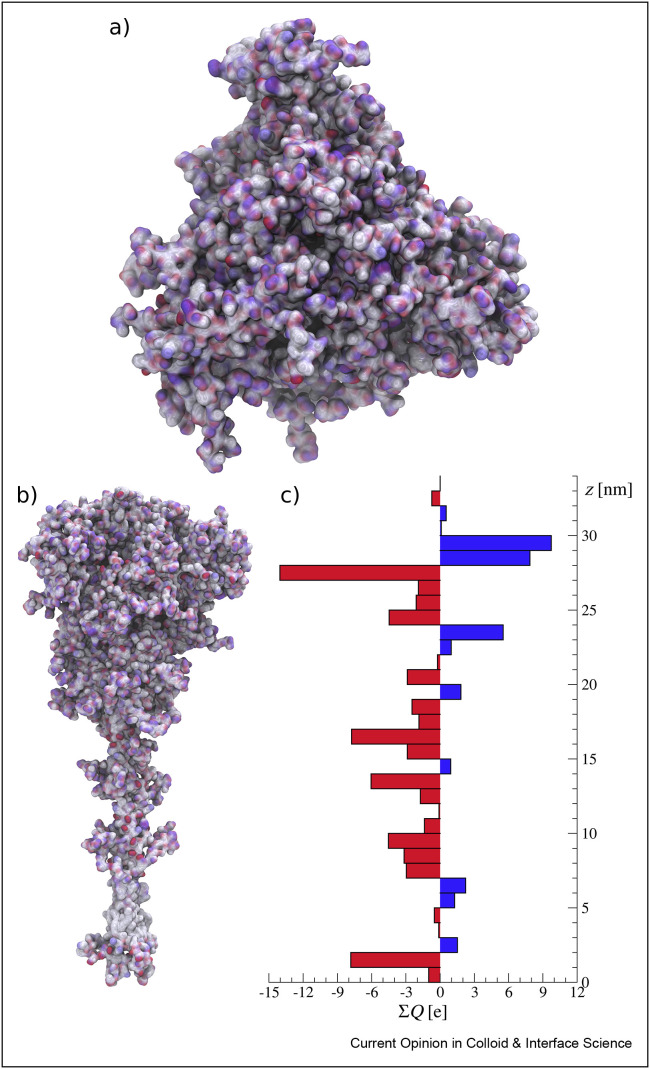
Figure 3.
Charge distribution of the SARS-CoV-2 S glycoprotein, (a) top view, (b) side view, and (c) the charge distribution in the direction z, perpendicular to the virus surface. The ΣQ denotes the total electrostatic (partial) charge in 1-nm thick slice. Red and blue colors denote negative and positive charges, respectively. Snapshots were made based on molecular coordinates provided in the study by Casalino et al. [22].
Using 15 nm as the plausible hydrodynamic diameter of the spike protein molecule, one can calculate from Eq. (1) that its diffusion coefficient in aqueous solutions is equal to 3.3 × 10− 1 1 m2 s−1 (at the temperature of 298 K). Given that the experimental measurements using conventional techniques, such as dynamic light scattering (DLS), are impractical, this estimate of the diffusion coefficient of the protein molecule may prove useful. Analogously, considering that the top cross section of the molecule resembles a triangle of which the side length equals 15 and 16 nm for the closed and open conformations, respectively, one can calculate that the surface area is equal to 97 and 110 nm2, respectively. This parameter is useful to predict the maximum coverage of protein molecules adsorbed on abiotic surfaces.
Physicochemical characteristics (nominal charge and charge distribution)
Charge distribution within protein molecules and their surfaces is a key factor in understanding their adsorption phenomena, which are often governed by electrostatics interactions [28]. It should be noted that because of the heterogeneous charge distribution, proteins with negative net charge can efficiently adsorb on negatively charged surfaces [28]. Therefore, the knowledge of charge distribution on SARS-CoV-2 S protein is of immense importance for prediction of its interaction with various surfaces. In Figure 3(a) and (b), the charge distribution over the spike protein in a closed state viewed from the top and side, respectively, is shown. In Figure 3(c), the distribution of partial charges along the protein determined within 1-nm thick slabs is shown. The overall nominal charge of the protein, determined from PROPKA for pH 7.4 [22,27], was equal to −32 e. However, based on Figure 3(c), one can clearly see the uneven charge distribution across the molecule. The top part is positively charged, whereas the rest of the molecule is negatively charged. One should also note that in physiological conditions, that is, characterized by 0.15 M NaCl concentration, the charge screening is efficient, so the effective electrostatic interaction range is of the order of a nanometer. Considering this and the fact that the spike protein molecules are oriented with the top part exposed to the bulk solution, one can deduce that the effective zeta potential of the virion should be positive, which can enhance its affinity to model surfaces used in experiments, such as mica [29], and to typical fomite surfaces, for example, silica, cellulose fibers, and so on [30, 31, 32, 33], which are often negatively charged. On the other hand, the middle and lower parts of the protein are negatively charged, which ensures its stabilization and prevent cluster formation, as shown experimentally in the study by Turoňová et al. [15].
Beside the charge distribution at physiological conditions (pH 7.4), it is interesting to know how the change in pH can influence the spike protein charge. It was shown that stability and activity of SARS-CoV-2 are significantly suppressed at pH <5 and pH >9 [34]. Interestingly, similar observations were previously reported for the SARS-CoV virus [35]. Recently, Zhou et al. [36] reported that at endosomal pH 4.0–5.5, the spike evades potentially neutralizing antibody through a pH-dependent mechanism of conformational masking.
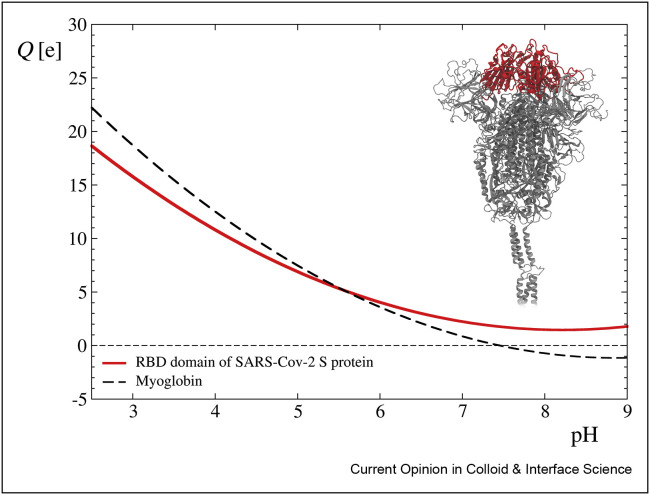
The dependence of the nominal (titrable) charge of protein molecules can be conveniently calculated using the PROPKA algorithm [27]. In Figure 4, we have presented the nominal charge of the receptor-binding domain (RBD) of the spike protein (PDB: 7BZ5) [37] (highlighted red in the inset) as a function of pH [38]. As can be seen, the nominal charge of the RBD domain remains positive for the entire pH range up to 9 that agrees with the charge distribution at pH 7.4 presented in Figure 3(c). Interestingly, the myoglobin molecule exhibits analogous dependence of the nominal charge for acidic and neutral pH range. Considering this analogy, one can calculate the electric potential distribution applying the method previously developed for myoglobin [38] exploiting the nonlinear Poisson–Boltzmann model, where the formation of two electric double layers within and outside the molecule was considered. The electric potential at the shear plane, experimentally accessible via electrophoretic mobility measurements [38], affects the RBD interactions (and in consequence the entire virion) with receptors and abiotic surfaces. This suggests that results of experimental measurements performed for myoglobin [38], especially the dependence of the zeta potential on pH, ionic strength, and temperature, can serve as useful reference data for predicting the attachment of the virion to receptors and abiotic surfaces.
Figure 4.
The nominal charge of the RBD of SARS-CoV-2 S protein [37] (highlighted red in the inset) versus pH, determined using the PROPKA algorithm. The black dashed line denotes data of nominal charge versus pH for the myoglobin molecule taken from the study by Batys et al. [38].
Attachment of spike protein to surfaces
Molecular dynamics and free energy modeling can also be used to derive quantitative information about the spike protein binding to the ACE2 receptor. To increase the efficiency of such calculations, only small fragments of the protein are considered, usually the RBD or the receptor-binding motif. In the study by Wang et al. [39], the dynamic trajectory method was applied to calculate the binding free energy. It is confirmed in this way that the binding strength of the SARS-CoV-2 RBD to the receptor is 10–15 times larger than that of the SARS-CoV RBD, which may contribute to increased infection rate. Interestingly, it is argued that the difference is due to enhanced electrostatic complementarity of the SARS-CoV-2 RBD to the ACE2 caused by the mutation in the hydrophobic residue and the removal of four proline residues. However, one should mention that in these calculations, the influence of ionic strength, pH, and electric potential appearing due to double-layer formation and specific ion adsorption were not considered.
The influence of ionic strength varied by NaCl between 0.15 and 1 M on the free energy of SARS-CoV-2 RBD binding to the ACE2 receptor was studied by Silva De Souza et al. [40] by molecular dynamics modeling and experimentally by the surface plasmon resonance (SPR) measurements. Using the free energy, it is predicted that the dissociation constant is equal to 7 × 10−10, 2.9 × 10−8, and 1 × 10−9 M for NaCl concentration equal to 0.15, 0.5, and 1 M, respectively. On the other hand, the experimental measurements showed that the RBD dissociation constant monotonically increases with NaCl concentration from 1.1 × 10−9 to 8 × 10−9 M for 0.15 and 1 M, respectively. Given that the dissociation constant assumes very low values, it is concluded that the increase in the electrolyte concentration does not break the SARS-CoV-2 RBD/hACE2 complex. However, the influence of pH and the electric potential distribution were not considered in these calculations.
Until recently, few theoretical investigations were devoted to the important issue of the spike protein interaction with abiotic surfaces. One of the exemptions represents the study by Malaspina and Faraudo [41], where the all-atom molecular dynamics (MD) modeling was used to determine the interactions of the SARS-CoV-2 spike protein with cellulose (assumed to be capable of forming strong hydrogen bonding) and graphite, a hydrophobic material. The protein with total charge of −23 e (corresponding to pH 7) was inserted in a water droplet, whereas the substrate was neutral and not hydrated. The system was neutralized by the 23 Na+ ions, which in result gives a very low ionic strength. The progress of the protein attachment (with the RBD directed toward the substrate surface) was monitored as a function of time by the number of residues in contact with the surface and the rms factor. It is confirmed that in the case of graphite, significant deformation of the protein appeared that decreased its binding strength.
One should underline that such molecular modeling is a powerful tool capable of resolving the structure–function relationship, which is often beyond the scope of experiments, and there is a need for particular effort in this direction.
Adsorption of S protein at surfaces
Few experimental investigations focused on quantifying the adsorption of the RBD fragment or the whole SARS-CoV-2 spike protein on abiotic surfaces were performed. This is mainly caused by a limited availability of the protein (usually produced in a recombinant form) which renders the commonly used methods such as the ellipsometry, reflectometry, SPR, or quartz crystal microbalance rather impractical, especially under flow conditions. Even simple physicochemical characteristics of protein molecules themselves by the DLS, which yields the diffusion coefficient (hydrodynamic diameter), and by the micro-electrophoretic techniques (laser Doppler velocimetry) are not performed because they require mg quantities of the solute. However, as shown in the study by Adamczyk et al. [25], an attractive alternative represents the AFM measurements that can be used for imaging single protein molecules on sufficiently smooth substrates, preferably mica or silica, yielding directly their surface concentration as a function of time. Using the high-speed AFM technique, quasi-continuous measurements of protein adsorption kinetics become feasible [29]. It is interesting to mention that under the diffusion transport applied in such measurements, the relaxation time of the protein monolayer formation is equal to [28].
| (4) |
where Γ mx is the protein monolayer coverage, D s is the protein diffusion coefficient, and c b is the protein mass concentration in the bulk.
One can predict from Eq. (4) that the monolayer formation time is inversely proportional to ; therefore, it attains large values for lower bulk protein concentration, allowing for thorough kinetic studies using AFM. For the spike protein where D s = 3.3 × 10−11 m2 s−1, assuming Γ mx = 0.7 mg m−2 and c b = 5 mg L−1, one can calculate from Eq. (2) that t m = 480 s.
The high-speed in situ AFM measurements of the spike protein adsorption kinetics on alumina and titania substrates were performed by Xin et al. [29] in NaCl and KCl solutions of various concentrations at pH 7.0–7.5. The coverage of the protein was determined via advanced image analysis software. It is shown that the characteristic time of monolayer formation on titania for pure water was equal to ca. 530 s, which agrees with the abovementioned estimate. However, the adsorption kinetics was much slower for the 0.165 M NaCl concentration, especially for the alumina substrate where the monolayer formation time increased to 4000 s. As argued in the study by Wasilewska et al. [42], where an analogous effect was observed for the human serum albumin adsorption on a silica substrate, the increased adsorption time can be attributed to the protein aggregation at such a large electrolyte concentration. Nevertheless, the results reported in the study by Xin et al. [29] unequivocally confirmed a significant adsorption of the spike protein on the oxide surfaces, which are negatively charged at pH of 7.5. This suggests that the protein exhibited a positive electrokinetic charge, at least on the RBD domain that qualitatively agrees with the theoretically predicted charge distribution (see Figure 4). However, because of the lack of the information about the zeta potential of the S protein and the substrates, a more quantitative analysis of these results obtained in the study by Xin et al. [29] is not feasible.
Another method requiring minor quantities of protein consists in controlled adsorption of proteins on carrier particles of nanometer to micrometer size range [25]. This leads to corona formation, mimicking in principle, inactive virion structure [43,44]. A considerable advantage of this method is that the monolayer formation time is considerably shorter than planar substrate adsorption even for dilute protein solutions. This relaxation time can be calculated from the formula
| (5) |
where is the maximum volume fraction of nanoparticles in the suspension, is the particle density, c m is the particle concentration in the suspension, d m is the particle diameter, and is the effective diffusion coefficient of the protein molecule relative to the particle (which is characterized by the diffusion coefficient equal to D m). One can calculate from Eq. (5) that for the spike protein molecule where D s = 3.3 × 10−11 m2 s−1 adsorbing on 100 nm in diameter gold particles (characterized by ρ m = 1.93 × 104 kg m−3) of the concentration c m = 100 mg L−1, the monolayer formation time is equal to 1.9 s, whereas for microparticle polymer carriers characterized by d m = 8 × 10−7 m (ρ m = 1.05 × 103 kg m−3) and for c m = 100 mg L−1, one obtains t m = 19 s.
Spike protein adsorption experiments on gold particles of the diameter varied between 10 and 100 nm were performed in the study by Yokoyama and Ichiki [44] applying the SPR method for a broad range of pH varied between 3 and 9.7. A significant attachment of the protein molecules was observed for the entire particle size range with the coverage quantified assuming a prolate spheroid shape of the protein molecule with the axis lengths equal to 14.5 and 16 nm. These values agree with the dimensions of the spike protein at the top shown in Figure 2. However, because the zeta potential of the gold particles (presumably negative for this pH range) and the protein were not determined or estimated, a quantitative analysis of these results is not feasible.
Considering these experimental data, one can argue that to unequivocally elucidate the spike protein adsorption, mechanisms on fomite surfaces thorough experimental measurements are needed. They should be performed for well-characterized systems using a combination of complementary methods, for example, the DLS, laser Doppler velocimetry, and concentration depletion methods as analogously to previously performed investigations for globular proteins [15,28].
Concluding remarks
Basic physicochemical data collected for the SARS-CoV-2 virion in this work can be used for predicting its transfer and attachment to abiotic surfaces. It is argued that in analogy to colloid particles, the overall virion attachment rate is controlled by its transfer through the bulk phase, usually through air or aqueous media in experimental investigations. However, the maximum number of attached virions is governed by its affinity to a substrate, which is controlled by the corona of spike proteins. Therefore, quantitative information about the spike protein, especially its interactions with various substrates, is of primary importance.
The analysis of published data derived from advanced molecular dynamic modeling and experimentally from cryo-EM confirmed that the charge distribution over the spike molecule is heterogeneous with the RBD positively charged for a broad range of pH. It is suggested that such a charge distribution pattern enhances the virion binding strength to receptors and abiotic surfaces, usually negatively charged.
One can also expect, given the risks involved with intact virion investigations, that attention in future works should be focused on elucidating mechanisms of the spike protein attachment to various substrates. Such experiments should be performed for substrates well-characterized in respect to chemical composition, surface morphology, and charge distribution, preferably applying a combination of complementary methods. Experiments involving carrier nanoparticles seem to be especially attractive, because of a fast kinetics and much lower protein consumption compared to flat substrate adsorption. In addition, the orientation of the spike protein molecules and its density in coronas formed in this way can be controlled by pH and the surface charge of carrier particles modified by macroion adsorption.
As far as theoretical investigations are concerned, one may suggest that future studies should be focused on elucidating the role of electric double layers, specific ion adsorption, and local pH changes in spike protein adsorption at various substrates.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was partially supported by the statutory activity of the Institute of Catalysis and Surface Chemistry, Polish Academy of Sciences. This research was supported in part by PLGrid Infrastructure.
This review comes from a themed issue on Hot Topic: COVID-19
Edited by Reinhard Miller and Libero Liggieri
References
- 1.Belosi F., Conte M., Gianelle V., Santachiara G., Contini D. On the concentration of SARS-CoV-2 in outdoor air and the interaction with pre-existing atmospheric particles. Environ Res. 2021;193:110603. doi: 10.1016/j.envres.2020.110603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noorimotlagh Z., Jaafarzadeh N., Martínez S.S., Mirzaee S.A. A systematic review of possible airborne transmission of the COVID-19 virus (SARS-CoV-2) in the indoor air environment. Environ Res. 2021;193:110612. doi: 10.1016/j.envres.2020.110612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poon W.C.K., Brown A.T., Direito S.O.L., Hodgson D.J.M., Le Nagard L., Lips A., et al. Soft matter science and the COVID-19 pandemic. Soft Matter. 2020;16:8310–8324. doi: 10.1039/d0sm01223h. [DOI] [PubMed] [Google Scholar]
- Duval J.F.L., van Leeuwen H.P., Norde W., Town R.M. Chemodynamic features of nanoparticles: application to understanding the dynamic life cycle of SARS-CoV-2 in aerosols and aqueous biointerfacial zones. Adv Colloid Interface Sci. 2021;290:102400. doi: 10.1016/j.cis.2021.102400. [DOI] [PMC free article] [PubMed] [Google Scholar]; A quantitative physicochemical concept of virus transmission via airborne particulate matter and aerosols droplets was elaborated and thoroughly discussed.
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (80- ) 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]; The structural analysis of the SARS-CoV-2 virion attachment to the human ACE2 receptor.
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marquès M., Domingo J.L. Contamination of inert surfaces by SARS-CoV-2: persistence, stability and infectivity. A review. Environ Res. 2021;193:110559. doi: 10.1016/j.envres.2020.110559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sellaoui L., Badawi M., Monari A., Tatarchuk T., Jemli S., Luiz Dotto G., et al. Make it clean, make it safe: a review on virus elimination via adsorption. Chem Eng J. 2021;412:128682. doi: 10.1016/j.cej.2021.128682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reina G., Peng S., Jacquemin L., Andrade A.F., Bianco A. Hard nanomaterials in time of viral pandemics. ACS Nano. 2020;14:9364–9388. doi: 10.1021/acsnano.0c04117. [DOI] [PubMed] [Google Scholar]
- 10.Chung Y.H., Beiss V., Fiering S.N., Steinmetz N.F. Covid-19 vaccine frontrunners and their nanotechnology design. ACS Nano. 2020;14:12522–12537. doi: 10.1021/acsnano.0c07197. [DOI] [PubMed] [Google Scholar]
- 11.Neuman B.W., Buchmeier M.J. Supramolecular architecture of the coronavirus particle. Adv Virus Res. 2016;96:1–27. doi: 10.1016/bs.aivir.2016.08.005. Academic Press Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner A., Autin L., Fuentes D., Maritan M., Barad B.A., Medina M., et al. CellPAINT: turnkey illustration of molecular cell biology. Front Bioinforma. 2021;1:660936. doi: 10.3389/fbinf.2021.660936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoňová B., Sikora M., Schürmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science (80- ) 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]; The SARS-CoV-2 virion structure, dimensions, the number of spike proteins and their distribution over the membrane, their size and orientations were determined by cryo-EM.
- 16.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; The SARS-CoV-2 virion dimensions, the number of spike proteins, their orientation angle distribution, size and shape were determined by cryo-EM.
- Kiss B., Kis Z., Pályi B., Kellermayer M.S.Z. Topography, spike dynamics, and nanomechanics of individual native SARS-CoV-2 virions. Nano Lett. 2021 doi: 10.1021/acs.nanolett.0c04465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun. 2020;11:1–10. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roselli R.J., Diller K.R. Springer; New York: 2011. Biotransport: principles and applications. [DOI] [Google Scholar]
- Casalino L., Gaieb Z., Goldsmith J.A., Hjorth C.K., Dommer A.C., Harbison A.M., et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent Sci. 2020 doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]; The SARS-CoV-2 S protein molecular structure derived from all-atom molecular dynamics simulations.
- 23.Fischer H., Polikarpov I., Craievich A.F. Average protein density is a molecular-weight-dependent function. Protein Sci. 2009;13:2825–2828. doi: 10.1110/ps.04688204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh Chaudhuri R., Paria S. Core/shell nanoparticles: classes, properties, synthesis mechanisms, characterization, and applications. Chem Rev. 2012;112:2373–2433. doi: 10.1021/cr100449n. [DOI] [PubMed] [Google Scholar]
- Adamczyk Z., Nattich-Rak M., Dąbkowska M., Kujda-Kruk M. Albumin adsorption at solid substrates: a quest for a unified approach. J Colloid Interface Sci. 2018;514:769–790. doi: 10.1016/j.jcis.2017.11.083. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]; The SARS-CoV-2 virion recognition of ACE2 receptor is experimentally determined yielding binding constants, antibody neotralisation of the S protein by antibodies is theoretically analyzed.
- 27.Olsson M.H.M., SØndergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical p K a predictions. J Chem Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 28.Adamczyk Z. Protein adsorption: a quest for a universal mechanism. Curr Opin Colloid Interface Sci. 2019;41:50–65. doi: 10.1016/j.cocis.2018.11.004. [DOI] [Google Scholar]
- Xin Y., Grundmeier G., Keller A. Adsorption of SARS-CoV-2 spike protein S1 at oxide surfaces studied by high-speed atomic force microscopy. Adv NanoBiomed Res. 2021;1:2000024. doi: 10.1002/anbr.202000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellezza F., Cipiciani A., Latterini L., Posati T., Sassi P. Structure and catalytic behavior of myoglobin adsorbed onto nanosized hydrotalcites. Langmuir. 2009;25:10918–10924. doi: 10.1021/la901448a. [DOI] [PubMed] [Google Scholar]
- 31.Lee J.G., Lannigan K., Shelton W.A., Meissner J., Bharti B. Adsorption of myoglobin and corona formation on silica nanoparticles. Langmuir. 2020;36:14157–14165. doi: 10.1021/acs.langmuir.0c01613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Żeliszewska P., Bratek-Skicki A., Adamczyk Z., Cieśla M. Human fibrinogen adsorption on positively charged latex particles. Langmuir. 2014;30:11165–11174. doi: 10.1021/la5025668. [DOI] [PubMed] [Google Scholar]
- 33.Wasilewska M., Adamczyk Z., Sadowska M., Boulmedais F., Cieśla M. Mechanisms of fibrinogen adsorption on silica sensors at various pHs: experiments and theoretical modeling. Langmuir. 2019;35:11275–11284. doi: 10.1021/acs.langmuir.9b01341. [DOI] [PubMed] [Google Scholar]
- 34.Chan K.H., Sridhar S., Zhang R.R., Chu H., Fung A.Y.F., Chan G., et al. Factors affecting stability and infectivity of SARS-CoV-2. J Hosp Infect. 2020;106:226–231. doi: 10.1016/j.jhin.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darnell M.E.R., Subbarao K., Feinstone S.M., Taylor D.R. Inactivation of the coronavirus that induces severe acute respiratory syndrome, SARS-CoV. J Virol Methods. 2004;121:85–91. doi: 10.1016/j.jviromet.2004.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou T., Tsybovsky Y., Olia A.S., Gorman J., Rapp M.A., Cerutti G., et al. A pH-dependent switch mediates conformational masking of SARS-CoV-2 spike. BioRxiv Prepr Serv Biol. 2020 doi: 10.1101/2020.07.04.187989. 2020.07.04.187989. [DOI] [Google Scholar]
- 37.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science (80- ) 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batys P., Nattich-Rak M., Adamczyk Z. Myoglobin molecule charging in electrolyte solutions. Phys Chem Chem Phys. 2020;22:26764–26775. doi: 10.1039/d0cp03771k. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu M., Gao J. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc Natl Acad Sci U S A. 2020;117:13967–13974. doi: 10.1073/pnas.2008209117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva De Souza A., Rivera J.D., Almeida V.M., Ge P., De Souza R.F., Farah C.S., et al. Molecular dynamics reveals complex compensatory effects of ionic strength on the severe acute respiratory syndrome coronavirus 2 spike/human angiotensin-converting enzyme 2 interaction. J Phys Chem Lett. 2020;11:10446–10453. doi: 10.1021/acs.jpclett.0c02602. [DOI] [PubMed] [Google Scholar]
- Malaspina D.C., Faraudo J. Computer simulations of the interaction between SARS-CoV-2 spike glycoprotein and different surfaces. Biointerphases. 2020;15 doi: 10.1116/6.0000502. [DOI] [PubMed] [Google Scholar]
- 42.Wasilewska M., Adamczyk Z., Pomorska A., Nattich-Rak M., Sadowska M. Human serum albumin adsorption kinetics on silica: influence of protein solution stability. Langmuir. 2019;35:2639–2648. doi: 10.1021/acs.langmuir.8b03266. [DOI] [PubMed] [Google Scholar]
- 43.Gorshkov K., Susumu K., Chen J., Xu M., Pradhan M., Zhu W., et al. Quantum dot-conjugated SARS-CoV-2 spike pseudo-virions enable tracking of angiotensin converting enzyme 2 binding and endocytosis. ACS Nano. 2020;14:12234–12247. doi: 10.1021/acsnano.0c05975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yokoyama K., Ichiki A. Nano-size dependence in the adsorption by the SARS-CoV-2 spike protein over gold colloid. Colloids Surf A Physicochem Eng Asp. 2021;615:126275. doi: 10.1016/j.colsurfa.2021.126275. [DOI] [PMC free article] [PubMed] [Google Scholar]