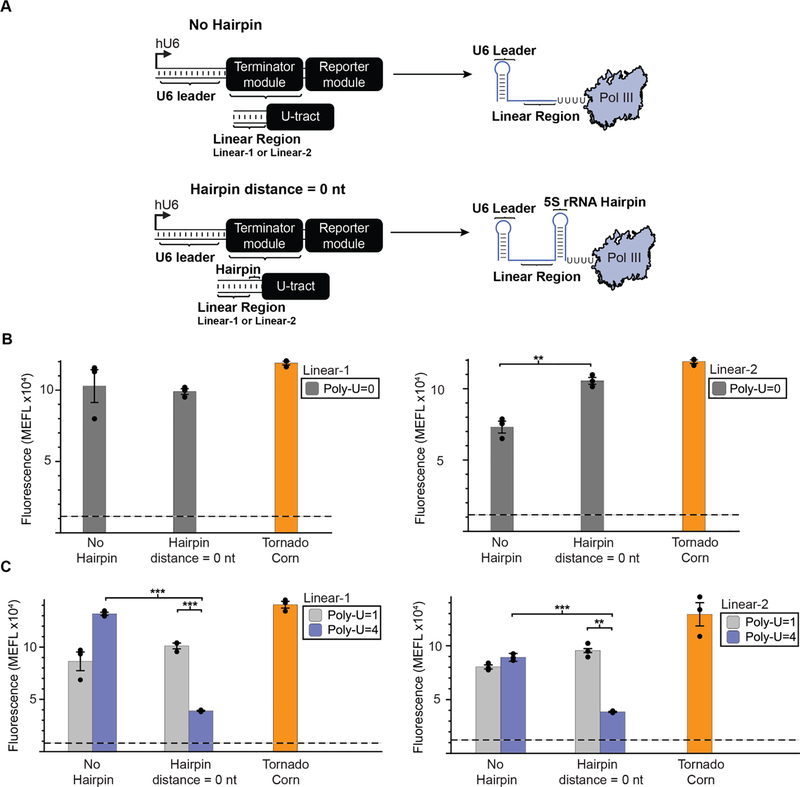
Figure 3: Upstream RNA secondary structure enhances termination as a function of poly-U tract length.
A) A schematic depicting the positioning of an RNA secondary structure element immediately upstream of the poly-U tract within the Terminator module. The secondary structure utilized is a 23 nt portion of the 5S ribosomal RNA (rRNA) predicted to fold into a 9 bp hairpin following previous studies of Pol III termination in vitro. This structure was either omitted (No Hairpin) or positioned after the linear sequence and immediately upstream of the poly-U tract (Hairpin distance = 0 nt). B) Impacts of upstream RNA structure to termination was evaluated for both the Linear-1 (left) and Linear-2 (right) sequence contexts when the poly-U tract is completely removed. C ) Contribution of upstream RNA structure to termination was evaluated for both the Linear-1 (left) and Linear-2 (right) sequence contexts, when the poly-U tract is changed from 1 to 4 uracils. Colored bars represent the average of 3 biological replicates with individual points plotted as circles. Error bars represent the S.E.M. The dashed line represents the average of three replicates for the vector-only control, and the grey horizontal bar represents the S.E.M. of the vector-only control, as reported in Fig. 2C. Statistical significance of the indicated comparisons (brackets) was measured using one-tailed heteroscedastic Welch’s t-tests followed by the Benjamini-Hochberg procedure with a false discovery rate cutoff of 0.05 (Supplementary Table 2). * = p < 0.05, ** = p< 0.01, *** = p < 0.001.