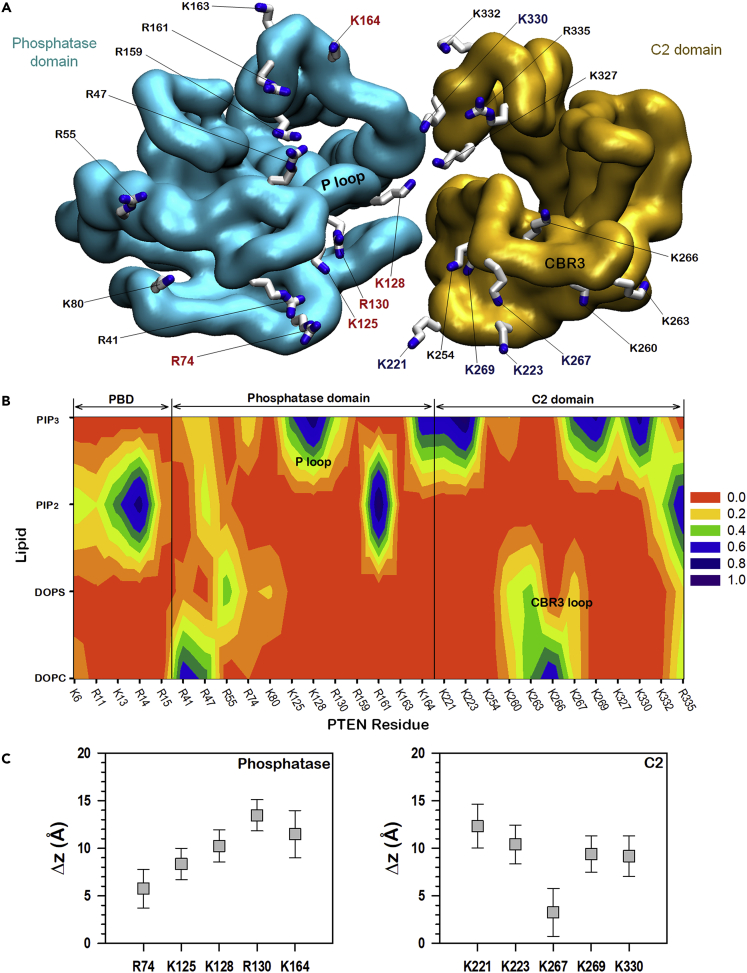
Figure 4.
Salt bridge interactions of PTEN
(A) Mapping of the basic residues on the membrane-binding surface of the phosphatase and C2 domains. PIP3-favored residues in the phosphatase domain are colored red and those in the C2 domain are colored blue.
(B) Contour map of the probabilities of salt bridge formations between the key basic residues at the membrane-binding interface and the lipids. Salt bridge is calculated between the nitrogen atoms in the side chains of basic residues and the oxygen atoms in the phosphate group of all lipids and the inositol head group of phosphoinositides with the cutoff distance of 3.2 Å.
(C) Averaged deviations of the amide nitrogen in the side chains of Arg and Lys residues from the bilayer surface for the PIP3-favored residues in the phosphatase (left panel) and C2 (right panel) domains. Error bars denote standard deviation.