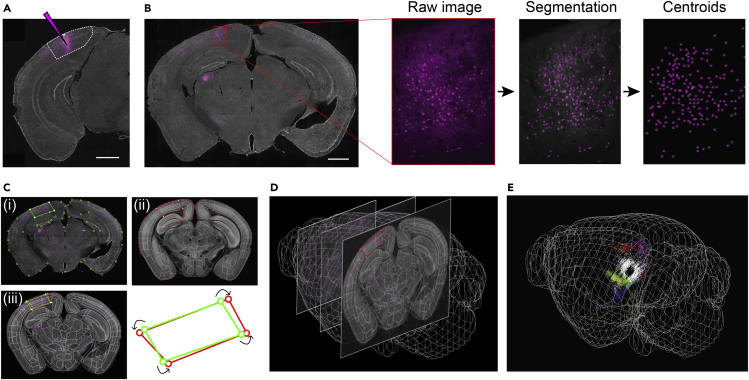
Figure 1.
Software pipeline for mapping cell body locations to brain regions through alignment with Allen CCF
(A) Example injection site of CTB in V1. White dotted line denotes V1 boundary based on the Allen Reference Atlas (ARA). Scale bar: 1 mm.
(B) Demonstration of the steps used to detect labeled cells: cells were segmented from the raw image and their centroids were extracted. Scale bar: 1 mm.
(C) Images of brain slices were registered to the ARA using SHARP-Track. We illustrate an example of this procedure, where transformation points on DAPI image ([i], green) were transformed to fit a reference slice (from ARA) transformation points ([ii], red). Overlay of (i) and (ii) shown in (iii).
(D) The registration was carried out across all brain slices along the anterior-posterior axis.
(E) An example visualization of cell body locations on the 3D brain model with cells color-coded by area identity (see also Figures S1, S2 and Video S1).