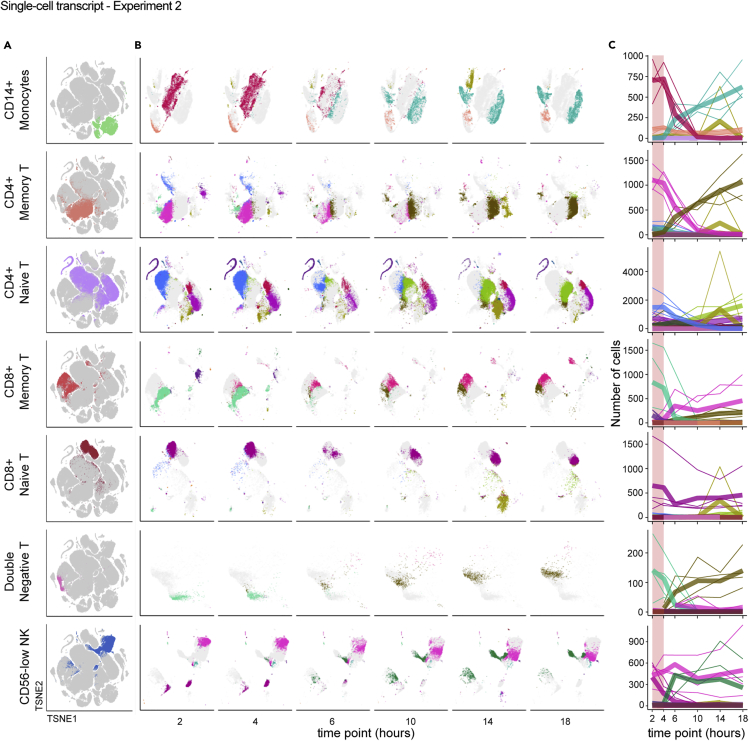
Figure 5.
Individual cell types undergo distinct but analogous paths away from the native transcriptional state over time
Single-cell RNA-sequencing data from Experiment 2 were analyzed as in Figure 4 and the STAR Methods.
(A and B) Data were isolated based on reference-based cell type, shown on the global tSNE map (A), and displayed separately based on time point using the same tSNE coordinates (B). Colors in (A) are assigned by cell-type label and are arbitrary. Each color in (B) represents a different Louvain cluster.
(C) The number of cells occupying each cluster was enumerated and the counts (y axis) plotted per cell type relative to the time after blood draw (x axis) for each donor. Colors match the Louvain cluster colors in (B). Thicker lines represent the mean for all donors and thinner lines represent single donors. The 2–4 hour range highlighted in pink shows a period of apparent stability prior to more extreme divergence in cluster occupancy after 4 hours.