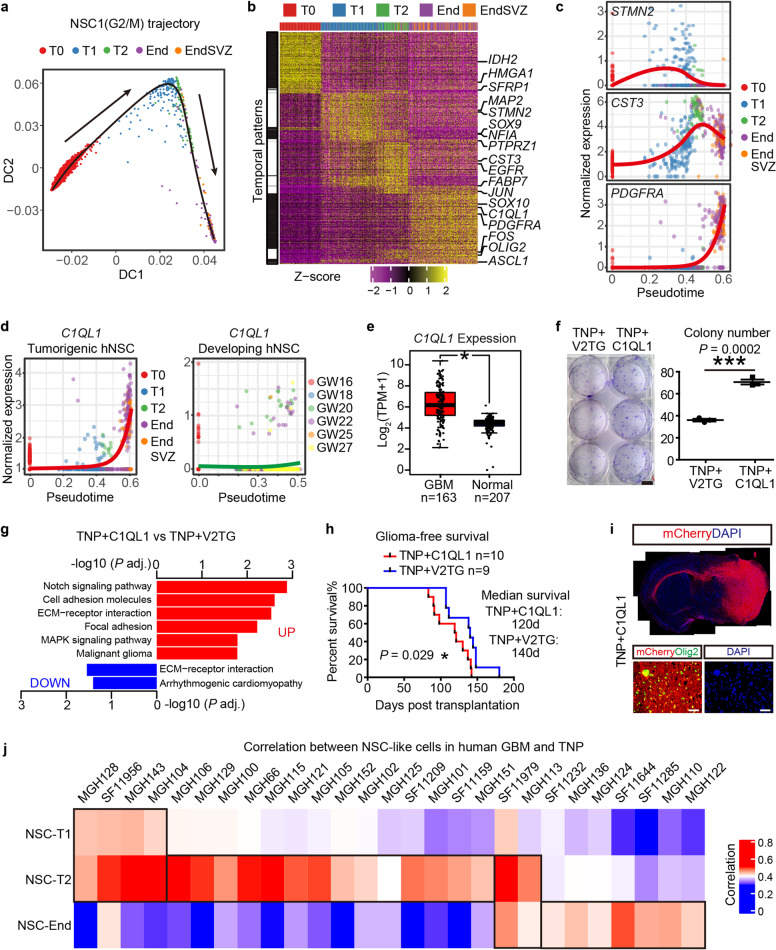
Fig. 6. Pseudotime analyses identify stage-specific regulatory programs.
a Diffusion map for NSC1 (G2/M), cells colored by stage. b Heatmap of gene expression cascades along the pseudotime of NSC1. Columns, pseudotime-ordered cells. Rows, genes. Colors in the top row indicate the actual sample time of each cell. Genes were clustered by their temporal expression patterns (left bars). c Trajectory plots of representative genes in (b) visualized by normalized expression. d Trajectory plots of C1QL1 in tumorigenic TNP hNSCs (left) and embryonic hNSCs during human hippocampal development (right). e The log2-transformed TPM of C1QL1 from TCGA GBM and matched GTEx normal brain RNA-seq. P < 0.01, one-way ANOVA test. f Left, colony formation assays of TNP + C1QL1 hNSCs and TNP + V2TG hNSCs. Scale bars, 1 cm. Right, the total number of colonies. n = 3 for each group. P values, Student’s t-test. g Gene ontology enrichment analyses of DEGs in TNP + C1QL1 vs TNP + V2TG hNSCs showing representative terms and adjusted P values (–log10). h The brain-tumor-free survival curves of TNP + C1QL1 and TNP + V2TG mice. n, the number of samples; P value, Log-rank (Mantel-cox) test. i IF for mCherry/DAPI and mCherry/OLIG2 in TNP + C1QL1 brains at the end stage. Scale bars, 1 mm (upper panels) and 50 μm (lower panels). j Heatmap of the Pearson correlation coefficients between NSC-like cells in human GBM samples (columns) and TNP samples at different stages (rows) based on their expression of DEGs in NSC1.