Summary
Clinically important broadly reactive B cells evolve during multiple infections, with B cells re-activated after secondary infection differing from B cells activated after a primary infection. Here we studied CD27highCD38high plasmablasts from patients with a primary or secondary dengue virus infection. Three transcriptionally and functionally distinct clusters were identified. The largest cluster 0/1 was plasma cell-related, with cells coding for serotype cross-reactive antibodies of the IgG1 isotype, consistent with memory B cell activation during an extrafollicular response. Cells in clusters 2 and 3 expressed low levels of antibody genes and high levels of genes associated with oxidative phosphorylation, EIF2 pathway, and mitochondrial dysfunction. Clusters 2 and 3 showed a transcriptional footprint of T cell help, in line with activation from naive B cells or memory B cells. Our results contribute to the understanding of the parallel B cell activation events that occur in humans after natural primary and secondary infection.
Subject areas: Immunology, Cell biology, Functional aspects of cell biology, Systems biology
Graphical abstract
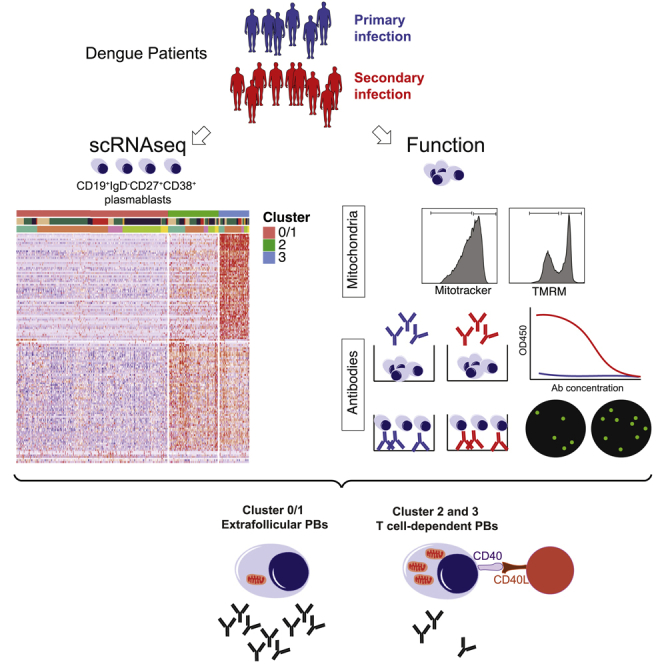
Highlights
-
•
CD27highCD38high plasmablasts from patients with dengue form three clusters
-
•
Clusters 0/1 express high levels, and more dengue-specific, antibodies
-
•
Clusters 0/1 are reminiscent of extrafollicular activation
-
•
Clusters 2 and 3 are metabolically active with an expression footprint of T cell help
Immunology; Cell biology; Functional aspects of cell biology; Systems biology
Introduction
Early activation of B cells and their differentiation into antibody-secreting plasmablasts is an integral part of a protective anti-viral response. Besides the B cell receptor (BCR)-mediated, antigen-specific activation, B cells can also be activated non-specifically by Toll-like receptor (TLR) ligands or by type I interferon (Bernasconi et al., 2003; Joo et al., 2012). Peak frequencies of plasmablasts are detected in the blood of infected patients approximately 7 days after infection (Carter et al., 2017; Ellebedy et al., 2016; Lavinder et al., 2014). In vitro, memory B cells differentiate efficiently into antibody-secreting plasma cells in the presence of IL-2, IL-10, CD40L, and BCR engagement, whereas the differentiation of naive B cells stops at the blasting stage, without any antibody secretion (Arpin et al., 1997). Owing to higher TLR expression in memory B cells compared with naive B cells, TLR-mediated stimulation triggers differentiation of memory B cells, but not naive B cell, into antibody-secreting plasma cells (Bernasconi et al., 2003). Interaction with T cell help within secondary lymphoid organs facilitates the activation of naive B cells in vivo, which can lead to the formation of germinal centers for B cell maturation (Tas et al., 2016). Selected B cells are retained in lymphoid or non-lymphoid organs for prolonged periods of time and form part of the immune memory (Kurosaki et al., 2015). During a subsequent infection with the same virus or an antigenically related virus, previously expanded “primary” memory B cells can re-enter germinal centers for further maturation and selection to become “secondary” memory B cells. Overall, a B cell response comprises the formation of memory B cells and plasmablasts, whereby only the latter have the capacity to secrete antibodies. Depending on the context of the infection, plasmablasts can originate from activated primary memory B cells, from secondary memory B cells, and possibly from activated naive B cells that never entered a germinal center. However, no markers are known to differentiate plasmablasts with these different origins (Garcia-Bates et al., 2013; Leach et al., 2019; Mesin et al., 2016; Purtha et al., 2011; Simon-Loriere et al., 2017; Stamper and Wilson, 2017; Tas et al., 2016).
Although being a cell population that is incompletely understood in terms of origin and relatedness to memory B cells and long-lived plasma cells, plasmablasts have been leveraged to isolate virus-specific, therapeutic antibody candidates (Corti et al., 2011; Wilson and Andrews, 2012; Xu et al., 2017). If a particular plasmablast subset is anticipated to contain antibodies with more therapeutic value, e.g., representing a more selected, reactivated secondary response versus a less selected primary B cell response, the ability to selectively screen such plasmablasts could more efficiently uncover therapeutically relevant antibody candidates. Therefore, phenotypic or genetic markers to identify these subsets could prove useful for antibody discovery.
Dengue virus (DENV) infection results in the formation of huge numbers of plasmablasts 5–7 days after the onset of fever (Appanna et al., 2016; Wrammert et al., 2012). DENV has four serotypes, immunity against each of which only provides serotype-specific protection in the long term (John and Rathore, 2019). Multiple infections are common in endemic countries, including Singapore, where this study was conducted. The virus surface glycoprotein, called E protein, is 63%–77% conserved between serotypes (Xu et al., 2016). During each infection, memory B cells to conserved regions of the virus are re-activated, expand exponentially, and may accumulate additional mutations upon re-entry into germinal centers. A frequent class of memory-derived antibodies is that binding to the fusion loop (FL) of the virus, which is highly conserved across DENV serotypes and other flaviviruses (Alwis et al., 2014; Lai et al., 2013; Xu et al., 2016). However, whereas fusion-loop-specific antibodies can be neutralizing at high concentrations, they have been shown to enhance viral infection at limiting concentration and can contribute to disease enhancement during secondary infection (Katzelnick et al., 2017). In turn, serotype-specific antibodies, including antibodies binding to complex epitopes (CE) that are only present on virus particles but not on recombinantly expressed E protein monomers, can have high neutralizing capacity. Examples of such CE antibodies are those binding only to E protein dimers (Dejnirattisai et al., 2015; Durham et al., 2019), to the hinge-region of the E protein when present at a certain angle (Fibriansah et al., 2014), or across E protein dimers (Fibriansah et al., 2015). Some of these CE-specific antibodies also neutralize multiple or all DENV serotypes (Dejnirattisai et al., 2015; Durham et al., 2019; Meihui et al., 2017; Rouvinski et al., 2015).
In this study we isolated plasmablasts from patients with primary or secondary dengue infection and identified three transcriptionally distinct plasmablast clusters (clusters 0/1, 2, and 3) in all patients tested. A significant number of differentially expressed genes (DEGs) between clusters were associated with oxidative phosphorylation. More metabolically active plasmablasts in clusters 2 and 3 were enriched with CD47-expressing cells. Independently, the percentage of CD47+ plasmablasts tended to be higher in patients with primary compared with secondary infection. Plasmablasts in clusters 2 and 3 expressed low levels of antibody genes and were more similar to memory B cells, whereas plasmablasts in cluster 0/1 were transcriptionally more related to, but not identical, to plasma cells. Cells in clusters 2 and 3 also showed a transcriptional footprint indicative of T cell help. Our results contribute to the understanding of the multiple B cell activation events that occur after natural primary and secondary infection in humans. The gene signatures we document here could also help in stratifying vaccine responses in the context of an individual's immune history.
Results
Plasmablasts after dengue infection can be divided into transcriptionally distinct subsets
The plasmablast response in patients after viral infection peaks between 5 and 7 days after onset of fever (Appanna et al., 2016; Wrammert et al., 2012; Xu et al., 2012). To address the transcriptional diversity of plasmablasts, we analyzed single plasmablasts from five patients with dengue, one with a primary infection and four with secondary infections, between days 4 and 9 after onset of fever (Table 1). The transcriptomes of a total of 890 single plasmablasts (defined as CD19+IgD−CD27hiCD38hi) were sequenced using Smart-seq2 technology (Figure 1A). Four plasmablast clusters were identified by employing Seurat and Singular algorithms to analyze DEGs. This analysis illustrated the existence of two similar clusters 2 and 3, which were clearly distinct from clusters 0 and 1 (Figure 1B). Cluster 3 was most distinct from the other clusters in both tSNE and UMAP dimensionality reduction analysis (Figure S1A). The DEGs between clusters 0 and 1 were mostly lambda and kappa genes, respectively, showing that the main difference between these clusters was the light chain usage (Figure S1B). Owing to this, clusters 0 and 1 were treated as one cluster in the following analysis. The most significant pathways that differentiated cluster 2 and 3 from the others were (1) EIF2 signaling, (2) oxidative phosphorylation, and (3) mitochondrial dysfunction. Regulation of eIF4 and p70S6K signaling, mTOR signaling, interferon signaling, and antigen presentation pathway genes were uniquely associated with cluster 2 (Figure 1C). When plasmablasts from two samples with an early day 4 time point (LNA018, LNA006) (Table 1) were analyzed, again four clusters were detected, and the most distinct pathways were (1) oxidative phosphorylation (2) mitochondrial dysfunction, and (3) EIF2 signaling, that the plasmablast clusters were not time point dependent. Cluster 0/1 was the largest cluster in all patient samples and time points analyzed, except for LNA018 early time point (Figure S2A and B). The immunoglobulin isotype distribution for all clusters was dominated by IgG1, most likely because four of the five patients experienced a secondary infection. There were, however, a higher number of cells expressing IgA1 in cluster 2 compared with the other clusters (Figure S2C). These cells could represent steady-state circulating plasmablasts originating from mucosal tissues (Iversen et al., 2017; Mei et al., 2009), although the mucosa-derived steady-state population that can be detected in any healthy individual was described to be dominated by the IgA2 isotype (Benckert et al., 2011; Spencer and Sollid, 2016).
Table 1.
Patient characteristics and assays performed with each blood sample
| Patient ID | Infection | Serotype | Sample time pointa | RNA-seq technology used | Total mAbs | DENV-binding mAb | B-cell fluorospot (Yes/No) |
Mitochondrial dye assay (Yes/No) | RT-PCR | ELISA (plasmablast supernatant) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CD47+/CD47- | IgG/A/M/neg | ||||||||||
| E1481DK2 | Secondary | DENV2 | 4–7 | Smart-seq2 | 0 | 0 | No | No | No | No | No |
| LNA002 | Primary | DENV2 | 6 | Smart-seq2 | 19 | 1 | No | No | No | No | No |
| LNA006 | Secondary | DENV2 | 5 | Smart-seq2 | 20 | 15 | No | No | No | No | No |
| 9 | Smart-seq2 | 39 | 25 | No | Yes | No | Yes | No | |||
| LNA018 | Secondary | DENV1 | 4 | Smart-seq2 | 10 | 6 | No | No | No | No | No |
| 6 | Smart-seq2 | 33 | 28 | Yes | No | Yes | No | No | |||
| LNA036 | Secondary | DENV2 | 4 | Smart-seq2 | 0 | 0 | No | No | No | No | No |
| 6 | Smart-seq2 | 28 | 22 | No | Yes | No | Yes | Yes | |||
| LNA014 | Primary | DENV1 | 9 | 10x Genomics | NA | NA | No | No | No | No | No |
| LNA023 | Secondary | DENV1 | 8 | 10x Genomics | NA | NA | Yes | No | No | No | No |
| LNA022 | Primary | DENV1 | 8 | NA | NA | NA | Yes | No | Yes | No | No |
| LNA049 | Primary | Unclear | 8 | NA | NA | NA | Yes | Yes | Yes | Yes | No |
| LNA034 | Primary | DENV1 | 8 | NA | NA | NA | Yes | Yes | Yes | Yes | No |
| LNA046 | Primary | Unclear | 7 | NA | NA | NA | Yes | No | Yes | No | No |
| LNA038 | Secondary | DENV2 | 7 | NA | NA | NA | Yes | Yes | Yes | Yes | No |
| LNA066 | Secondary | Suspected DENV2 | 8 | NA | NA | NA | Yes | No | Yes | No | No |
| LNA054 | Secondary | DENV1 | 7 | Smart-seq2 (CD47+/CD47-) | NA | NA | Yes | No | Yes | No | No |
| LNA063 | Secondary | DENV1 | 7 | Smart-seq2 (CD47+/CD47-) | NA | NA | Yes | No | Yes | No | No |
| LNA059 | Primary | DENV1 | 6 | NA | NA | NA | Yes | No | Yes | No | No |
| LNA064 | Primary | DENV1 | 6 | NA | NA | NA | Yes | No | Yes | No | No |
| LNA009 | Primary | Suspected DENV2 | 7 | NA | NA | NA | No | No | No | Yes | No |
| LNA010 | Primary | DENV1 | 7 | NA | NA | NA | No | Yes | No | Yes | No |
| LNA013 | Primary | DENV1 | 8 | NA | NA | NA | No | No | No | No | Yes |
| LNA041 | Primary | Unclear | 8 | NA | NA | NA | No | Yes | No | Yes | No |
| LNA043 | Primary | DENV2 | 6 | NA | NA | NA | No | No | No | No | Yes |
| LNA005 | Secondary | DENV3 | 8 | NA | NA | NA | No | No | No | Yes | No |
| LNA008 | Secondary | DENV2 | 6 | NA | NA | NA | No | No | No | No | Yes |
| LNA062 | Secondary | DENV3 | 8 | NA | NA | NA | No | Yes | No | Yes | No |
NA: not applicable
In days after fever onset.
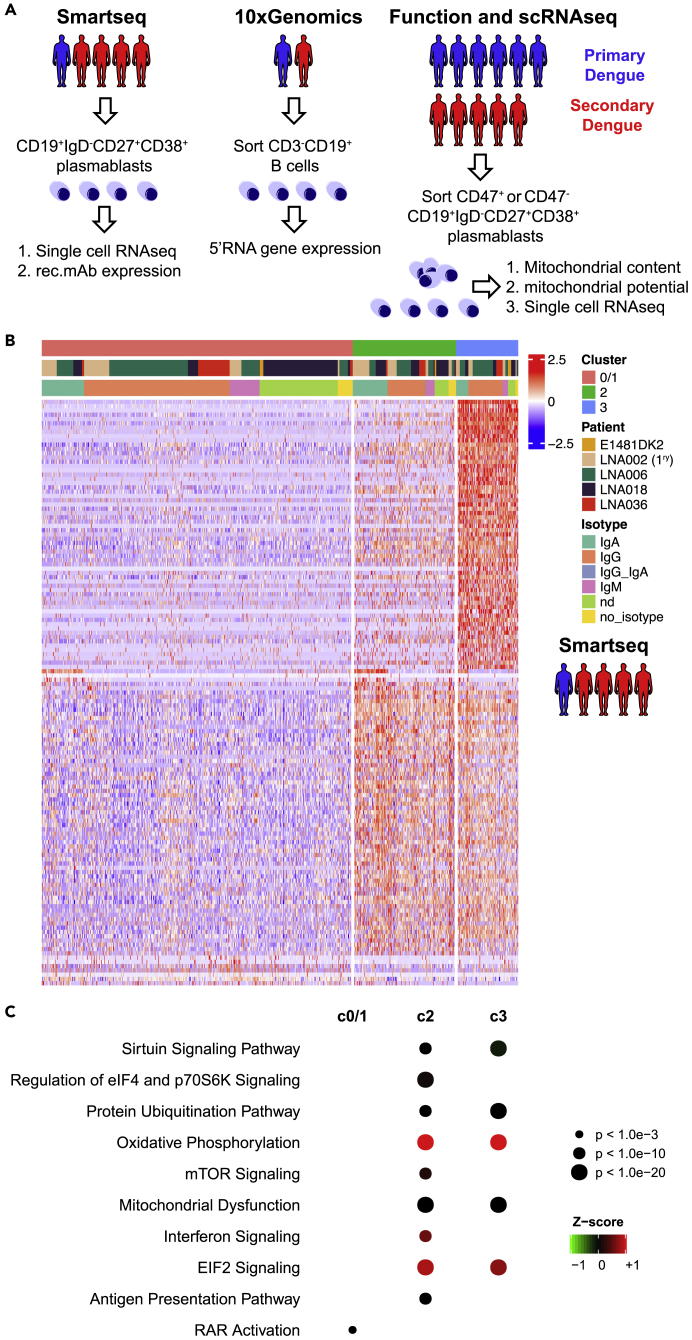
Figure 1.
Plasmablasts from patients with dengue show three distinct clusters based on their transcriptome
(A) Experimental setup: Plasmablasts (CD19+IgD−CD27+CD38+) were sorted from the blood of five dengue-infected patients 4–9 days after fever onset for single-cell RNA-seq analysis (Smart-seq2). Monoclonal antibodies (mAbs) were expressed recombinantly from 149 randomly selected cells. 10x Genomics 5′ sequencing was conducted from CD3−CD19+ cells sorted from two dengue-infected patients. CD47+ or CD47- plasmablasts (CD19+IgD−CD27+CD38+) from 11 dengue-infected patients were stained with MitoTracker Green (mitochondrial content) and TMRM (mitochondrial potential) and were sorted for RNA sequencing (two secondary patients).
(B) Heatmap representing supervised hierarchical clustering of 890 single plasmablasts from 5 patients using the 400 most variable genes as identified by Singular. 3 clusters were identified (0/1, 2 and 3). For each cell (column) the patient of origin and the isotype of the antibody expressed is indicated by the colored bar.
(C) Pathways that differentiate the 3 clusters (0/1, 2 and 3). DEGs from each cluster compared with the two others were used for Ingenuity Pathway Analysis.
The top DEGs for cluster 0/1 compared with other clusters were IGLC2 followed by 12 variable region genes and IGJ, indicating that these cells' transcriptional activity is dominated by the production of antibodies. In contrast, apart from the top DEG IGHA2 and the fourth-most significant DEG IGHG3, no additional Ig genes were among the top 100 DEGs for clusters 2. No Ig transcripts were among the top 100 DEGs of cluster 3 (Table S1). Clusters 2 and 3 therefore seem to express, and possibly secrete, less antibodies compared with cluster 0/1.
In summary, plasmablasts after dengue infection showed three transcriptionally distinct phenotypes, differing most significantly in the expression of antibody genes and genes associated with mitochondrial processes involved in energy generation, regulation of eIF4 and p70S6K signaling, and antigen presentation.
Functional analysis of antibodies derived from cells with high and low metabolic activity
To study the function of antibodies produced by transcriptionally distinct plasmablasts, 149 of the 890 total plasmablasts were selected randomly for expression of monoclonal antibodies (mAbs) (Table 1). Only IgG-expressing B cells were included for recombinant mAb expression as human IgG1. In the secondary patients, 96 of 130 (71%) of the mAbs bound to DENV when tested in ELISA plates coated with virus particles from all four serotypes (Figure 2A). As expected, given the early time point in the disease, few IgG mAbs from the primary patient were specific for DENV (1 of 19 mAbs expressed and tested). In line with this, antibodies secreted from bulk-sorted plasmablasts (PBs) from two additional primary patients bound less to DENV compared with those from secondary patients (Figure S3). These data suggested that a majority of the activated IgG-expressing B cells with a plasmablast phenotype in flavivirus-naïve patients were activated non-specifically.
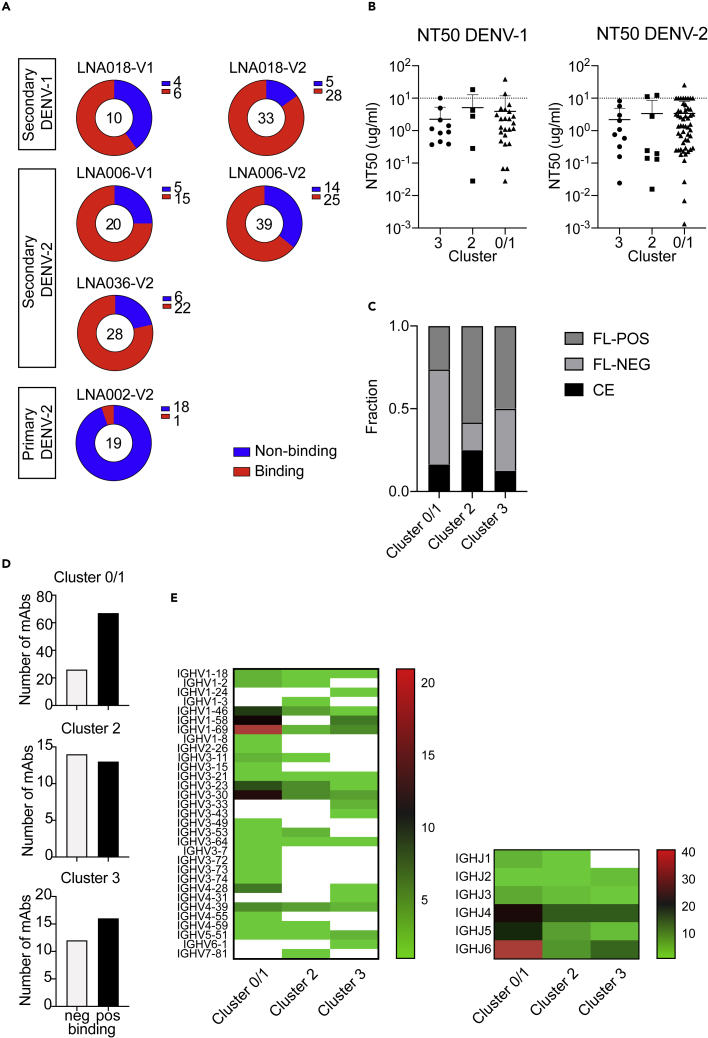
Figure 2.
Cluster 0/1 contains most DENV-specific cells and shows a distinct V and J gene usage
(A) Pie charts summarizing the number of mAbs expressed from each patient at visit 1 (V1) or visit 2 (V2) (see Table 1) and their ability to bind to DENV. Binding was determined by ELISA using PEG-DENV virus from the four serotypes. Binding was considered positive when mAbs recognized at least one serotype. A total of 149 mAbs were expressed and tested.
(B) DENV-1 (left) or DENV-2- (right) neutralizing titers (NT50) of individual mAbs grouped by cluster of origin. Each dot represents one mAb. The dotted line indicates the highest concentration tested. Bars indicate means and SD; no statistically significant difference between the three groups (Kruskal-Wallis test).
(C) Fraction of mAbs in each cluster (0/1, 2, or 3) binding to fusion loop (FL-POS, dark gray), to a complex epitope (CE, black), or to neither of these categories (FL-NEG, light gray).
(D) Bar charts representing the binding of individual mAbs according to the cluster (0/1, 2, or 3) of the plasmablast from which the mAb was cloned.
(E) IGHV gene usage (left) and IGHJ gene usage (right) in mAbs from the three clusters. White boxes indicate that the gene was not expressed by any of the mAbs contained in the cluster.
A larger fraction of the mAbs expressed from cluster 0/1 plasmablasts bound to DENV compared with mAbs from clusters 2 and 3 (Figure 2D). However, the neutralization capacity of all mAbs that bound to DENV was similar for all clusters for both DENV-1 and DENV-2 (Figure 2B), the serotypes prevalent in Singapore during the time of the study.
Further characterization by ELISA was conducted to test (1) whether antibodies bind to whole virus particles (UV-inactivated DENV particles) or to recombinant E protein monomer and (2) whether antibodies bind to three amino acids in the FL that were previously shown to be highly immunogenic, using E protein alanine substitution mutants (Dejnirattisai et al., 2015; Lai et al., 2013). The FL was of interest because it is a highly conserved region across all DENV serotypes and up to 80% of the IgG+ plasmablasts that are reactivated from memory B cells during secondary infection in patients can be specific to the FL (Appanna et al., 2016; Dejnirattisai et al., 2015). FL-specificity could therefore indicate memory B cell origin. Antibodies were grouped into FL-pos (loss of binding to G100A, W101A, G106A, and/or F108A E protein mutants), FL-neg, and CE-specificity. Clusters 2 and 3, which expressed few antibody genes and were metabolically more active, contained a larger proportion of FL-binding antibodies compared with cluster 0/1, although differences between clusters were not statistically significant (p value 0.06, chi-square test) (Figure 2C). Pathway analysis of the plasmablast transcriptome data based on the binding class of their respective antibodies showed that FL-specific cells (W101+F108-) expressed 15 genes that were clearly distinct compared with plasmablasts with other specificities (top five DEGs: MRPS34 SON, HCST, ATP5D, and COX7B, Figure S4A). The pathway associated with the 15 DEGs was highly dominated by energy metabolism, similar to the profile of clusters 2 and 3 (Figure S4B), providing further evidence that B cells specific for a conserved, highly immunogenic epitope were associated with these two clusters.
In line with distinct epitopes of mAbs from the individual clusters (Figure 2D), the clusters showed differential VDJ gene usage. Strikingly, cluster 0/1 was highly dominated by VH1-69 and VH1-58 gene usage combined with J6 gene usage (Figures 2E and S5A). This combination of variable gene usage was associated almost exclusively with FL-neg antibodies (Figure S5B).
Taken together, cluster 0/1 contained more virus-specific cells compared with clusters 2 and 3. Among DENV-binding antibodies, fusion-loop-specific antibodies were more abundant in clusters 2 and 3. Therefore, high metabolic activity in clusters 2 and 3 appeared to be associated (1) with activated specific B cells preferentially binding to a conserved and highly immunogenic epitope and (2) with activated unspecific B cells.
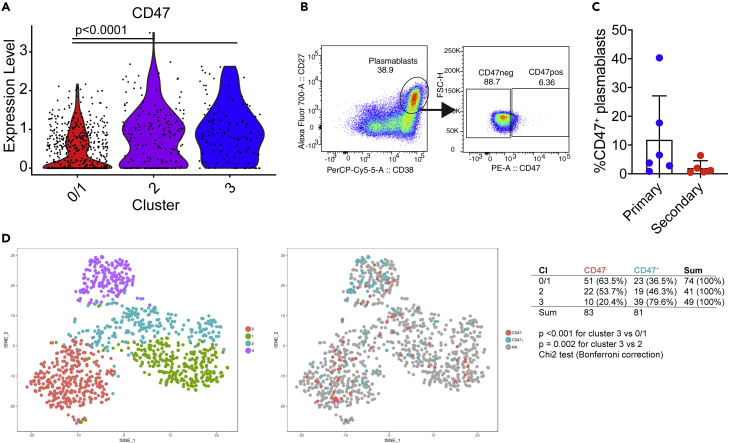
CD47 gene expression is higher in cluster 2 and 3 plasmablasts
To identify surface protein markers for flow cytometry-based sorting and functional analysis, we stained cells for a number of surface markers (Table 2) that were identified based on differential expression between cluster 3 and the other clusters. Cluster 3 was chosen based on its more distinct separation in cluster analysis compared with cluster 2 (Figure S1B). Markers were also selected based on high transcription level and based on the availability of antibodies to stain them. Among the markers tested, CD298 (ATP1B3), CD98 (SLC3A2), CD164, CD317 (BST2), CLEC2D, and CD48 were either not expressed on the surface of plasmablasts or uniform expression on all or a majority of plasmablasts was observed (Figures S6A and S6B). However, we identified CD47 as a marker with differential cell surface expression on a small subset of plasmablasts. CD47 gene expression was significantly upregulated in both clusters 2 and 3 compared with cluster 0/1 (Figure 3A). CD47 cell surface expression was low in general but higher in a fraction of cells (Figure 3B, gating strategy in Figure S6A). A total of 11 patients (6 primary and 5 secondary) were analyzed for CD47 expression on the surface of plasmablasts. Interestingly, some primary patients had very high proportions of CD47+ plasmablasts, which was not seen for secondary patients (Figure 3C). CD47 is expressed on various cell types and in various tissues. CD47 binds to signal regulatory protein alpha (SIRPα) expressed on phagocytic cells including macrophages and dendritic cells and negatively regulates phagocytosis (Barclay and Berg, 2014). To assess CD47 as a potential marker to identify cluster 2 and 3 cells, CD47-low and CD47-high plasmablasts from two additional secondary patients were sorted and analyzed using single-cell mRNA sequencing. The new data from 192 cells (96 cells CD47+ and 96 cells CD47-) were combined with the older dataset of 890 plasmablasts for tSNE clustering. The sorted CD47+ cells were significantly more abundant in cluster 3 and to a lesser extent in cluster 2 for both patients, validating CD47 as a marker to at least enrich for plasmablasts from these cluster (Figure 3D).
Table 2.
Genes upregulated in cluster 3 compared with other clusters tested for cell surface expression
| p_val | avg_logFC | pct.1 | pct.2 | p_val_adj | Plasmablast surface expressiona | |
|---|---|---|---|---|---|---|
| CD48 | 1.02E-06 | 0.015496825 | 0.879 | 0.674 | 0.020169 | Yes |
| CLEC2D | 7.83E-10 | 0.010398923 | 0.931 | 0.766 | 1.55E-05 | No |
| BST2 | 3.32E-11 | 0.009364097 | 0.862 | 0.643 | 6.56E-07 | Yes (very low) |
| CD164 | 2.65E-07 | 0.006051495 | 0.957 | 0.789 | 0.00525 | Yes |
| CD47 | 1.99E-11 | 0.011958774 | 0.879 | 0.659 | 3.93E-07 | Yes |
| SLC3A2 | 6.14E-12 | 0.009602143 | 0.724 | 0.386 | 1.21E-07 | Yes |
| ATP1B3 | 7.15E-25 | 0.015775382 | 0.828 | 0.49 | 1.41E-20 | Yes |
As measured by flow cytometry.
Figure 3.
CD47 gene and protein expression as a marker associated with clusters 2 and 3
(A) Gene expression level of CD47 in single plasmablasts from the three clusters (0/1, 2, and 3).
(B) Representative flow cytometry plot of the gating to identify surface CD47+ plasmablasts in one secondary patient (LNA054). Plasmablasts were gated as CD19+IgD−CD27+CD38+.
(C) Percentage of surface CD47+ plasmablasts in primary (n = 6, blue dots) and secondary (n = 5, red dots) patients at 6–8 days after fever. Data are combined from 3 independent experiments. Bars indicate means ± SD.
(D) Transcriptome data from single CD47+ or CD47- cells from two secondary patients were combined with the 890 cells originally analyzed (Figure 1). The left panel represents the 4 clusters obtained by Seurat (0, 1, 2 and 3). The right panel highlights CD47+ and CD47- plasmablasts in these clusters. The table summarizes the number of CD47- or CD47+ cells in each cluster. Chi-square test with Bonferroni correction was used to determine significant enrichment of CD47+ cells in cluster 3 compared with others.
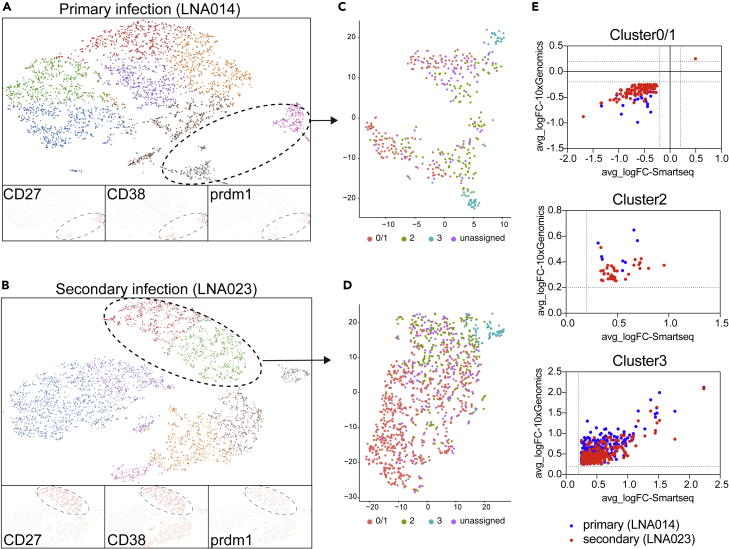
Plasmablasts from patients with primary versus secondary infection show distinct mitochondrial profiles that help characterize the nature of clusters 0/1 versus 2 and 3 cells
We first employed 10x Genomics-droplet-based transcriptomics as an independent technology to validate the findings and to assess the existence of plasmablast subsets in additional patients. Plasmablasts among total B cells were identified based on CD27, CD38, and PRDM1 expression (Figure 4A). In both a primary (LNA014) and a secondary patient (LNA023), plasmablasts formed sub-clusters when analyzed separately from other B cells (Figure 4B). The scPred algorithm was used to assign plasmablasts to the previously defined clusters (Figure 4B). For clusters 0/1 and 3, DEGs between 10x and Smart-seq technology correlated highly significantly. Few cells were assigned to cluster 2, making it difficult to determine the agreement between the two technologies for this cluster.
Figure 4.
Validation of PB clusters by 10x Genomics
(A and B) Graph-based representation of 5′ gene expression analysis (Loupe Cell Browser software) of B cells (CD3−CD19+) from primary patient LNA014 (A) and secondary patient LNA023 (B). Individual gene expression for CD27, CD38, and PRDM1, which were used to identify plasmablasts, are indicated.
(C and D) Assignment to Smart-seq2-defined clusters. Plasmablast clusters (circled populations) were extracted and assigned to clusters 0/1, 2, and 3 using scPred. tSNE representation of plasmablast clusters for the primary (C) and secondary patient (D), with assigned color-coded cluster identity.
(E) Correlations between average log fold change (avg_logFC) of up- or downregulated genes present in both Smart-seq2 and 10x Genomics datasets for clusters 0/1, 2, and 3. Only genes with an FC above/below +/− log0.2 were considered. Spearman correlation; Cluster 0/1: p < 0.0001 for LNA023, ns for LNA014, Cluster 2: ns, Cluster 3: p < 0.0001 for LNA023 and LNA014.
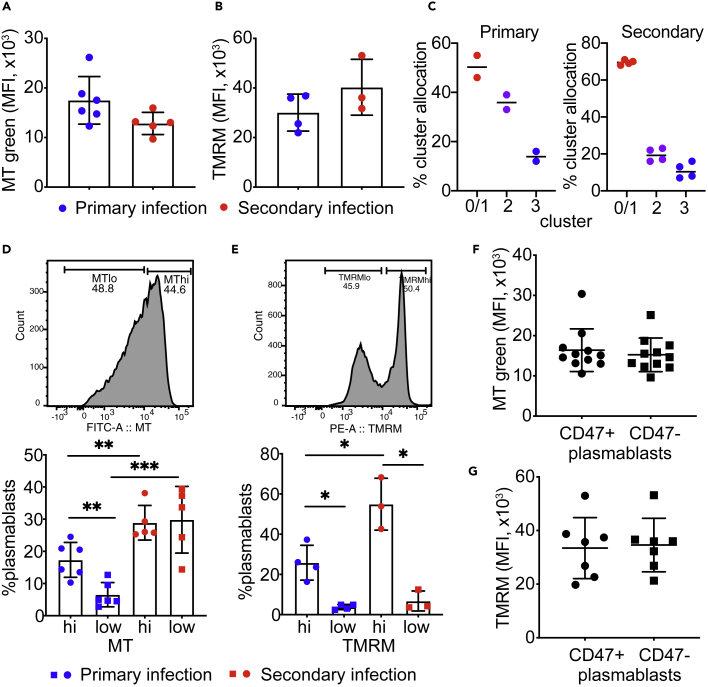
The results from the cluster analysis (Figure 1C) suggested an important role of mitochondrial activity, notably oxidative phosphorylation, in differentiating the clusters. To further understand the metabolic differences in PB subsets we conducted functional assays, assessing (1) the abundance of mitochondria by MitroTracker Green (MTgreen) staining and (2) the mitochondrial membrane potential by TMRM staining, which is a readout for active versus dysfunctional mitochondria.
PBs from primary patients tended to have higher MTgreen expression (mean fluorescence intensity [MFI]) compared with secondary cases (not statistically significant), whereas the opposite trend was seen for TMRM (Figures 5A and 5B). This suggested that primary patients' plasmablasts could contain more mitochondria. In line with this, primary patients showed relatively less cluster 0/1 and more cluster 2 (more metabolically active, Figure 1C) plasmablasts compared with secondary patients (Figure 5C). Although assessed in few patients, the ratio between the different clusters was surprisingly consistent. We next analyzed the percentage of plasmablasts that were MTgreen-low (MTlo) or MTgreen-high (MThi) and TMRM-low (TMRMlo) or TMRM-high (TMRMhi) as a fraction of CD19+ cells (gating strategy in Figure S7A). Significantly higher percentages of plasmablasts from primary patients fell into the MTgreen-high population compared with the MTgreen-low population, adding evidence for their higher mitochondria content. Secondary patients' plasmablasts had higher percentages of plasmablasts overall but showed similar percentages of MTgreen-high and -low cells (Figures 5D and 5E). Mitochondrial mass and mitochondrial function, as assessed by the MFI of MTgreen and TMRM, were not different between CD47+ and CD47- plasmasblasts (Figures 5F and 5G). Furthermore, sorted CD47+ and CD47- plasmablasts from primary or secondary patients secreted similar amounts of IgG (Figure S7B).
Figure 5.
Secondary infection plasmablasts have less mitochondrial content but higher mitochondrial potential
(A) Mean fluorescence intensity (MFI) of MitoTracker Green in plasmablasts (CD19+IgD−CD27+CD38+) from primary (n = 6, blue dots) and secondary patients (n = 5, red dots) at 6–8 days after fever.
(B) MFI of TMRM in plasmablasts (CD19+IgD−CD27+CD38+) from primary (n = 4, blue dots) and secondary patients (n = 3, red dots) 6–8 days after fever.
(C) Allocation of plasmablasts into clusters 0/1, 2, and 3 in primary and secondary patients, based on Smart-seq2 and 10x Genomics scRNA-seq data. Between 96 and 1,023 single cells per patient were used for this analysis. Patient E1481DK2 was excluded from the analysis because only 16 total cells were analyzed from this patient.
(D) Representative histogram of the MitoTracker Green expression level in plasmablasts (CD19+IgD−CD27+CD38+) from one patient (Top panel). Percentages of MTlo or MThi cells are represented in the bar chart for primary (n = 6, blue dots) and secondary (n = 5, red dots) patients at 6–8 days after fever.
(E) Representative histogram of the TMRM expression level in plasmablasts (CD19+IgD−CD27+CD38+) from one patient (top panel). Percentages of TMRMlo or TMRMhi cells are represented in the bar chart for primary (n = 4, blue dots) and secondary (n = 3, red dots) patients at 6–8 days after fever.
(F) MFI of MitoTracker Green in CD47+ or CD47- plasmablasts from 11 patients (6 primary and 5 secondary) at 6–8 days after fever.
(G) MFI of TMRM in CD47+ or CD47- plasmablasts from 7 patients (4 primary and 3 secondary) at 6–8 days after fever. Data in all panels are combined from 3 individual experiments. Means ± SD (A, B, D−G) or means (C) are indicated with bars. Statistics in (D) and (E): paired (PB from same donors) or unpaired (PB from different donors) t test. ∗∗∗p = 0.0006, ∗∗p = 0.0062 and 0.0064, ∗p = 0.01–0.015.
Taken together, the CD47 gene is expressed significantly higher in clusters 2 and 3 compared with cluster 0/1. Higher surface protein CD47 can also be used as a marker to enrich, but not to clearly separate, plasmablasts clusters 2 and 3 from cluster 0/1. Plasmablasts in secondary patients tended to have a lower mitochondrial mass and showed relatively less MThi cells compared with primary patients. A low mitochondrial mass is aligned with the transcriptional profile of cluster 0/1, which was not enriched with mitochondria-associated pathways (Figure 1C). Owing to the limitation of the low surface expression of CD47, MT staining could therefore provide an additional means to enrich for plasmablasts from clusters 0/1 versus clusters 2 and 3.
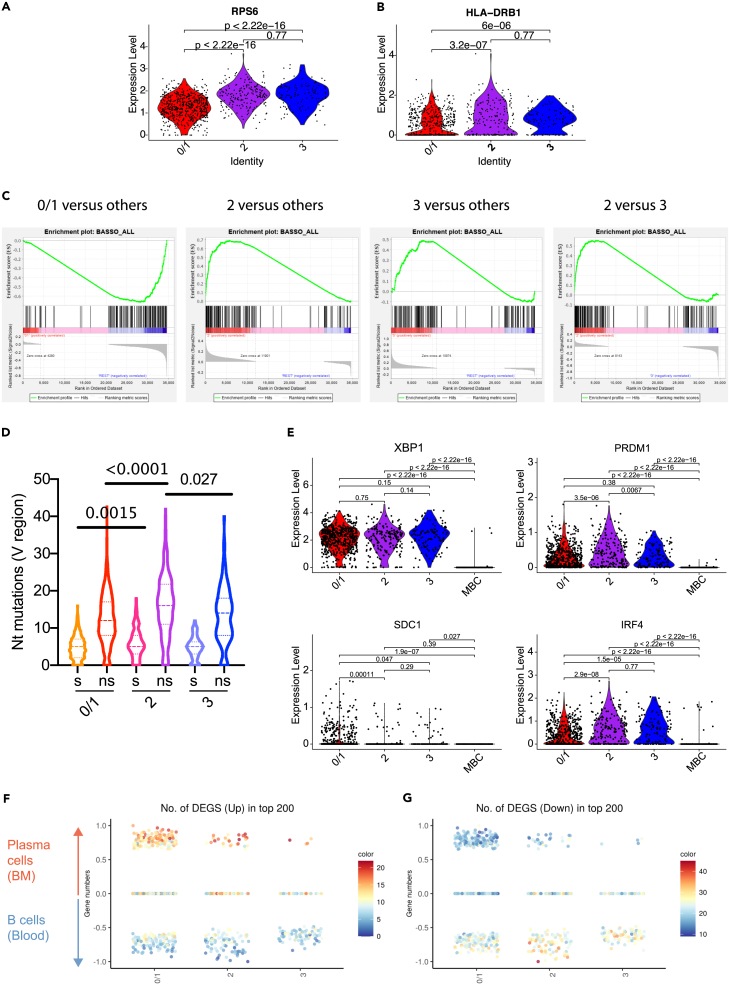
Plasmablasts in clusters 2 and 3 show footprints of T cell help-dependent activation
The differences in mitochondrial mass and metabolic activity for primary versus secondary patients (Figures 1C and 4) pointed to a different type of activation. Of note, genes associated with antigen presentation were significantly upregulated in cluster 2 (Figure 1C). We therefore hypothesized that differential involvement of T cell help during the activation of B cells contributed to the transcriptome clustering of plasmablasts. For reference, we first compared our clusters with the previously described transcriptional profiles of B cells activated in the presence of CD40L (Basso et al., 2004; Luo et al., 2018). RPS6, a gene downstream of CD40L (Luo et al., 2018), was expressed significantly higher in clusters 2 and 3 compared with cluster 1 (Figure 6A). In addition, HLA-DRB1, an MHC class II gene, was expressed significantly higher in clusters 2 and 3 (Figure 6B). A gene set enrichment analysis showed that clusters 2 and 3 were significantly enriched in genes downstream of CD40L activation (Figure 6C), adding further evidence for interaction with helper T cells in germinal centers or possibly in extrafollicular patches that have been described in the context of autoimmune and anti-pathogen responses in mice (Di Niro et al., 2015; William et al., 2002). In line with this, cells in cluster 2 showed significantly higher levels of silent and non-silent nucleotide mutations in the antibody variable regions compared with cluster 1 (Figure 6D). The number of mutations was similar for DENV-specific and non-specific mAbs (Figure S8). We speculate that non-specific cells were non-specifically activated memory B cells, or specifically activated naive or memory B cells that incorporated mutations that resulted in loss of binding.
Figure 6.
Plasmablasts from clusters 2 and 3 express genes that are in line with T cell help-mediated activation, whereas plasmablasts from cluster 1 show a plasma cell profile
(A) Expression level of RSP6 in single plasmablasts from clusters 0/1, 2, and 3.
(B) Expression level of HLA-DRB1 in single plasmablats from clusters 0/1, 2, and 3.
(C) GSEA plots of CD40L downstream genes enrichment in cluster 0/1 versus others, cluster 2 versus others, cluster 3 versus others, and cluster 2 versus cluster 3. Enrichment of 101 up-regulated and 63 down-regulated genes (gene sets from [Basso et al., 2004]) are represented.
(D) Level of silent (s) and non-silent (ns) nucleotides mutations in plasmablasts from cluster 0/1, cluster 2, and cluster 3. The median and quartiles are indicated with horizontal dotted lines. Statistics: Kruskal-Wallis test.
(E) Expression level of PRDM1, XBP1, SDC1, and IRF4 genes in single plasmablasts from clusters 0/1, 2, and 3, and in single memory B cells (MBCs) from the same dataset.
(F and G) Comparison of the level of expression of genes expressed by at least 10% of cells (n = 6,957 genes) with the dataset from (Gutiérrez et al., 2007) that established 146 up-regulated DEGs to identify plasma cells (in bone marrow) and 775 down-regulated DEGs to identify normal B lymphocytes (in blood). Single-cell gene expression in plasmablasts from individual cluster were defined as closer to “plasma cell” (between >0 and 1) or to “B lymphocytes” (between <0 and −1) or to none of them (0). Each dot indicates the enrichment score of a single plasmablast. The color coding represents the number of up- and downregulated genes, respectively, found in the top 200 most expressed genes for each cell. Statistics for gene expression comparisons between clusters in A,B and E: Wilcoxon test.
To assess a functionally important, potential difference in antibody secretion activity we assessed PRDM1 (Blimp-1) gene expression, which is required for high Ig production in antibody-secreting cells (Kallies et al., 2007). Clusters 2 and 3 expressed higher levels of blimp-1 (PRDM1) compared with cluster 0/1 (Figure 6E). This was unexpected given that cluster 0/1, but not clusters 2 and 3, showed high antibody gene expression levels. Another plasma cell marker, SDC1 (CD138), showed increased expression in cluster 0/1 compared with cluster 2 and/or 3; IRF4 showed decreased expression, whereas XBP1 expression was not significantly different. We also compared the clusters with previously published transcriptional data of sorted human bone marrow plasma cell and peripheral blood B cells from healthy donors, composed mostly of naive B cells (Gutiérrez et al., 2007). More plasmablasts in cluster 0/1 showed an expression profile similar to plasma cells, compared with plasmablasts in clusters 2 and 3, consistent with higher Ig gene expression in cluster 0/1 (Figures 6F and 6G).
These results confirmed the fundamentally different functions of phenotypically identical CD27highCD38high plasmablast clusters. Cluster 0/1 cells are functionally similar to antibody-secreting extrafollicular plasma cells that originate mostly from memory B cells based on higher percentage of specificity (Figure 2D). In contrast, cluster 2 and 3 cells appeared to be a mixed population of naive B cells activated with recent T cell help based on the CD40L signal footprint (Figure 6A), and of memory B cells activated in a non-antigen-specific manner. We base the latter observation on the finding that (1) clusters 2 and 3 contained less dengue-specific cells (Figure 1), (2) that cluster 2 showed a higher rate of non-synonymous mutations (Figure 6D), and (3) that MitoTracker-high cells, representative of more metabolically active clusters 2 and 3, were more abundant in primary patients (Figure 5).
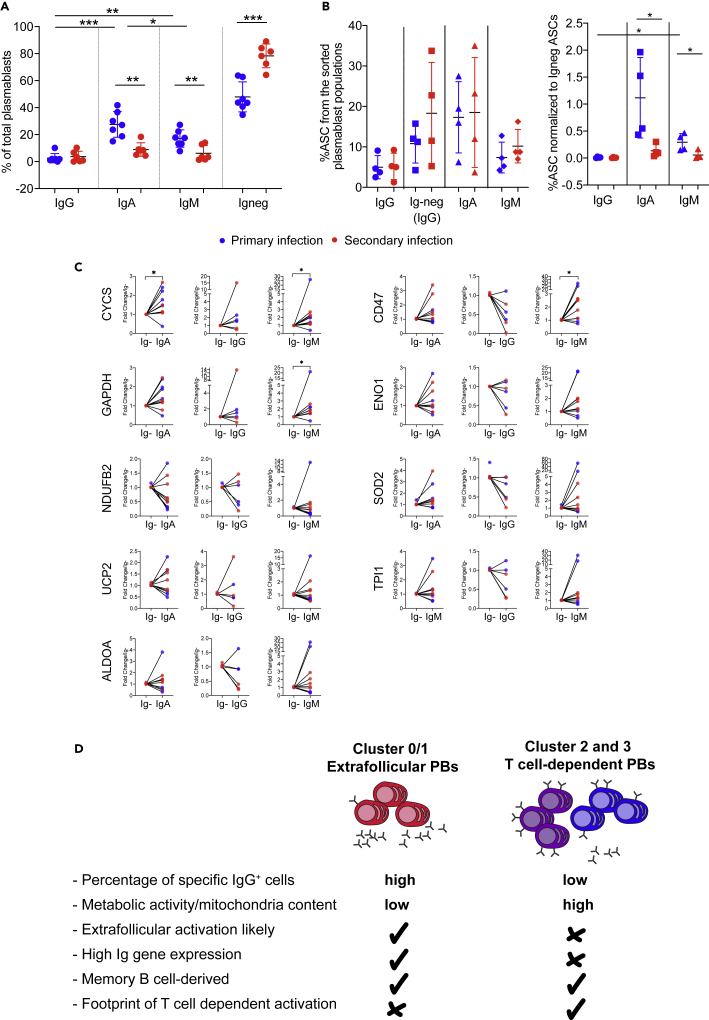
Association of metabolic activity with plasmablasts Ig secretion and isotype
To further understand the function of plasmablast clusters, and to build on the observation that clusters 0/1 expressed more Ig genes compared with clusters 2 and 3, we sorted plasmablasts based on isotype and surface expression of immunoglobulins (Igs). We then tested an association of surface Ig expression with metabolic activity based on gene expression defined in the previous paragraphs. IgA and IgM surface expression defined clearly distinct populations, whereas IgG surface expression was generally low (Figure S9), as described previously for plasma cells (Blanc et al., 2016; Pinto et al., 2013). Igneg plasmablasts were the largest fraction for both primary and secondary patients, whereas primary patients showed more IgM- and IgA-expressing cells (Figure 7A). When tested in an enzyme-linked immune absorbent spot assay after overnight incubation with R848 and IL2 to increase Ig secretion (Dorner et al., 2009), the Igneg fraction contained more IgG-producing cells compared with the IgG-pos fraction, showing that the Igneg fraction represented mostly IgG+ cells that had down-regulated BCR surface expression (Figure 7B). We next tested the expression of genes associated with the pathways that were significantly different between clusters 0/1 and 2/3 (Figure 1C). Cytochrome c (CYCS) expression was significantly higher in the IgA and IgM fraction compared with the Igneg fraction, and CD47 expression was higher in the IgM fraction (Figure 7C).
Figure 7.
IgM, IgA, and IgG-negative plasmablasts contain mostly cluster 0/1 cells
(A) Percentage of IgA+, IgM+, IgG+, and Ig− plasmablasts based on flow cytometry analysis (see gating strategy in Figure S9).
(B) ELISPOT-based number of antibody-secreting cells (ASCs) among the sorted populations (left panel), and the same data normalized to the number of IgG+ ASCs of the Igneg fraction per sorted sample (right panel). Bars in (A and B) indicate means ± SD. Paired (same donors) or unpaired (different donors) t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(C) Expression of metabolic genes found to be differentially expressed between cluster 0/1 and 2/3 in plasmablasts sorted and shown in (A) and (B). Wilcoxon matched-pairs signed rank test. ∗p < 0.05.
(D) Summary of characteristics for cluster 0/1, cluster 2, and 3 described in this study.
These data suggested that the Igneg fraction contained more cluster 0/1 cells compared with the IgA and IgM fractions, whereas the trend was less clear for surface IgG-positive cells, where less samples were available. Considering these data together with the single-cell RNA sequencing (scRNA-seq) clustering data (Figure 7D), we found that plasmablasts that have downregulated IgG surface expression secrete higher levels of antibodies after potential extrafollicular activation, as described for cluster 0/1 cells. In turn, plasmablasts with Ig surface expression secrete relatively less antibodies and are more likely to belong to clusters 2 and 3.
Discussion
Identification of plasmablasts by flow cytometry based on high surface expression on CD27 and CD38 is established in the field. Our study demonstrates that this phenotype comprises three transcriptionally distinct clusters. Cluster 0/1 (extrafollicular plasmablasts) expressed high levels of antibody genes and was transcriptionally closer but not identical to bone marrow plasma cells, based on cMAP analysis and based on the expression of classical plasma cell markers prdm1, SDC1, IRF4, and XBP1. Clusters 2 and 3 (T-cell-dependent plasmablasts) expressed few and very low levels of antibody genes, respectively, and showed a transcriptional profile of T-cell-dependent activation. Although no single surface marker tested here could clearly distinguish the clusters, CD47 was identified as a cell surface marker that could partially enrich for clusters 2 and 3 (Figure 3). Although higher metabolic activity was associated with clusters 2 and 3 based on gene expression, a direct link between surface CD47 expression and MitoTracker-high profile could not be established, possibly due to the low surface expression of CD47. A role for CD47 on plasmablasts has not been described. In plasma cells from patients with multiple myeloma, CD47 is upregulated compared with healthy plasma cells, and the function of CD47 was associated with cell activation and adhesion (Gutiérrez et al., 2007). Ligation of CD47 on vascular cells by its ligand thrombospondin-1 (TSP1) inhibits nitric oxide production (Isenberg et al., 2006). Ligation of CD47 by TSP1, which can be released from activated platelets during acute infection and inhibition of nitric oxide-related pathways in B cells, might be related to the high metabolic activity that was observed in clusters 2 and 3 (Lacza et al., 2009). Interestingly, long-lived plasma cells (LLPCs) have a higher maximal respiration capacity compared with short-lived plasma cells and use pyruvate import into mitochondria to sustain the high respiratory capacity (Lam et al., 2016). The “mitochondrial dysfunction” pathway of plasmablasts appears to suggest a limited lifespan. Alternatively, the higher number of mitochondrial genes could also reflect a higher number of mitochondria per cell as a consequence of a higher energy demand, in line with oxidative phosphorylation, which was also significantly enriched in clusters 2 and 3. It is also possible that only a portion of a clonally expanded B cell is activated and shows an activated blasting, i.e., plasmablast phenotype with a limited lifespan, whereas “sister” cells originating from the same B cell remain in the immune cell pool as memory B cells, thereby preserving a given specificity.
Functionally, more antibodies cloned and expressed from clusters 0/1 cells were antigen-specific and preferentially bound to CEs and non-FL epitopes. Antibodies cloned from clusters 2 and 3 were less likely to be specific, and if specific, were dominated by FL-binding antibodies. This was unexpected, as FL-specific cells appeared not to be re-activated memory B cells, as could have been expected due to the high conservation of the FL among flaviviruses. As T cell help is likely to have led to the activation of cells in clusters 2 and 3, this finding indicated that T cell help is readily available for FL-specific B cells, potentially explaining the immunodominance of this epitope (Angelo et al., 2017).
Our findings could be of importance for the discovery of therapeutic antibodies because potent cross-reactive antibodies are more likely to be affinity-matured and memory B cell-derived (de Alwis et al., 2012; Dejnirattisai et al., 2015; Meihui et al., 2017), and would therefore be more prevalent in cluster 0/1. Although no difference in neutralizing capacity was identified between antibodies isolated from plasmablasts from the different clusters, this could be explained by the rarity of potently cross-neutralizing antibodies in general.
It is curious that patients with dengue generate huge plasmablast responses even after a primary infection. We propose that, despite their identical phenotype based on high expression of surface markers CD27 and CD38, primary infection plasmablasts are functionally different from plasmablasts generated after secondary infection, particularly in terms of antibody-secreting capacity. Independent of primary or secondary infection, plasmablasts that are activated from naive B cells might not be efficient in secreting antibodies. Antibody secretion was not significantly different in CD47+ and CD47- cells (Figure S7B). However, because it was not possible to sort clean populations of cluster 3 and cluster 2 cells based on CD47 expression, their different antibody secretion capacity could not be confirmed. In addition to the limitations of CD47 as a surface biomarker, our analysis is more focused on secondary patients, particularly for scRNA-seq and mAb data, and more studies are needed to clearly dissect the extrafollicular versus T cell-dependent nature of plasmablasts after primary infection. In this context it would be interesting to study the response of children, who are more likely to experience a primary infection. The different expression profiles of clusters 0/1 and 2/3 could also point to a difference in plasmablast maturation stage, with clusters 2 and 3 representing recently activated, immature plasmablasts expressing less Ig genes and secreting less antibodies compared with cluster 0/1. The higher Ig mutation rate in clusters 2 and 3 does not support this explanation but does not exclude that at least part of the cluster 2 and 3 cells are immature plasmablasts.
Variable gene usage was surprisingly biased in cluster 0/1 versus clusters 2 and 3. The preference of heavy chain VH1-69 gene usage paired with J6 was almost exclusively found in FL-neg antibodies in cluster 0/1. Several potent cross- or broadly neutralizing dengue-, influenza-, and HIV-specific antibodies found in large screens and selected for potential therapy share VH1-69 and J6 usage (Chen et al., 2019). Many of these antibodies were highly mutated, in line with selection during multiple germinal center reactions. The cells identified here as cluster 0/1 plasmablasts contained such selected memory B cells that were likely activated in an extrafollicular response without T cell help.
It was unexpected that cluster 0/1 showed lower blimp-1 expression compared with clusters 2 and 3, despite the highest Ig gene expression. Possibly, the PRDM1 gene was downregulated after an initial upregulation. Alternatively, the low level of PRDM1 in all clusters could indicate that PBs differentiate independent of blimp-1, as shown in mice that expressed gfp in the locus of PRDM1 (Kallies et al., 2007). In this model, cells lacking blimp-1 cell were CD138 negative and could still secrete antibodies, although at a very low level. Bcl-6, a gene that is down-regulated upon B cell activation and differentiation into plasmablasts and plasma cells, was not differently expressed between the clusters.
Overall, we show a previously unappreciated diversity of phenotypically identical CD27hiCD38hi human plasmablasts after dengue infection. Cluster 0/1 plasmablasts represent classical antibody-secreting cells that are more likely to be re-activated memory B cells participating in an extrafollicular response. Clusters 2 and 3, which are enriched in CD47+ cells, represent naive or memory B cells that show characteristics of T cell-dependent activation. It will be interesting to validate these profiles in other human infections and to further establish phenotypic differences in B cell activation to guide the isolation of therapeutic antibodies and to better characterize natural and vaccine-induced responses in individuals with or without previous exposure to the same antigen.
Limitations of the study
Although we provide readouts including the cloning and testing of recombinant antibodies from plasmablasts and quantification of metabolic activity to assess the potential function of transcriptionally distinct plasmablast clusters, our conclusions are largely based on scRNA-seq data. The current lack of a clear surface marker to distinguish clusters 0/1, 2, and 3 limits the bulk sorting and further functional analysis of the plasmablast subsets.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Pacific blue-anti-human-CD19 (HIB19) | Biolegend | 302232 |
| PerCP-Cy5.5-anti-human-CD38 (HIT2) | BD Biosciences | 561106 |
| AF700-anti-human-CD27 (O323) | Biolegend | 302814 |
| PE-Cy7-anti-human-IgD (IA6-2) | Biolegend | 348210 |
| PE-anti-human-CD47 (REA220) | Miltenyi | 130-123-980 |
| FITC-anti-human-CD47 (REA220) | Miltenyi | 130-101-345 |
| APC-anti-human-IgA (IS11-8E10) | Miltenyi | 130-113-998 |
| Anti-human-Ig purified (polyclonal) | Thermofisher Scientific | H1700 |
| Anti-human-IgG AF488 (polyclonal) | Invitrogen | A11013 |
| Anti-human-IgA purified (polyclonal) | Jackson Immunoresearch | 109-005-011 |
| Anti-human-IgM Pacific blue (MHM-88) | Biolegend | 314513 |
| Anti-human-IgM APCCy7 (MHM-88) | Biolegend | 314520 |
| Anti-human-CD27 PE (M-T271) | BD Biosciences | 555441 |
| Anti-human-CD19 BV650 (HIB19) | Biolegend | 302237 |
| Anti-human-IgG BV510 (G18-145) | BD Biosciences | 563247 |
| Bacterial and virus strains | ||
| DENV-1 strain D1/SG/05K2916DK1/2005 | Environmental Health Institute EHI, Singapore | Genbank: EU081234.1 |
| DENV-2 strain DENV-2-TSV01 | Novartis Institute of Tropical Diseases, Singapore | Genbank: AY037116.1 |
| DENV-1 strain DENV-1-WestPac74 | Novartis Institute of Tropical Diseases, Singapore | Genbank: U88535.1 |
| Biological samples | ||
| Blood | Tan Tock Seng Hospital (TTSH), Singapore | NA |
| Chemicals, Peptides, and recombinant proteins | ||
| LIVE/DEAD stain | ThermoFisher Scientific | L34957 |
| Tetramethylrhodamine, methyl ester, Perchlorate | ThermoFisher Scientific | T668 |
| Mito Tracker™ Green | ThermoFisher Scientific | M7514 |
| TetraMethylBenzidine (TMB) | Sigma Aldrich (Merck) | 54,827-17-7 |
| Resiquimod (R848) | InVivogen | tltl-r848 |
| IL-2 | Prospecbio | CYT-209 |
| KLH | Sigma Aldrich (Merck) | 9013-72-3 |
| Critical commercial assays | ||
| SD BIOLINE dengue duo kit for NS1 detection | Abbott | 11FK46 |
| PanBio dengue IgG indirect ELISA | Abbott | 01PE30 |
| OneStep RT-PCR kit | Qiagen | 210210 |
| Protein G mag sepharose | Cytiva (formerly GE Healthcare) | 28951379 |
| Alexa Fluor™ 488 antibody labeling kit from | ThermoFisher Scientific | A20181 |
| Chromium single cell 5′ library and gel bead kit | Research Instrument – 10X genomics | 1000014 |
| Chromium single cell V(D)J enrichment kit, human B cell | Research Instrument – 10X genomics | 1000016 |
| Chromium single cell 3’/5′ library construction kit | Research Instrument – 10X genomics | 1000020 |
| Chromium i7 multiplex kit | Research Instrument – 10X genomics | 120262 |
| Chromium single cell a chip kit | Research Instrument – 10X genomics | 1000009 |
| PicoPure™ RNA isolation kit | ThermoFisher Scientific | KIT0204 |
| Nextera XT DNA library preparation kit | Illumina Singapore | FC-131-1096 |
| Nextera XT index kit v2 | Illumina Singapore | FC-131-2001 FC-131-2002 FC-131-2003 FC-131-2004 |
| Experimental models: Cell lines | ||
| Vero cells | ATCC | ATCC® CCL-81™ |
| HEK293-6E | National Research Council of Canada | NRC file 11,565 |
| Oligonucleotides | ||
| GAACATGGRACAAYTGCAACYAT | Integrated DNA Technologies | DENV-1 D1MGBEn469s-forward |
| CCGTAGTCDGTCAGCTGTATTTCA | Integrated DNA Technologies | DENV-1 D1MGBEn536r-reverse |
| ACACCACAGAGTTCCATCACAGA | Integrated DNA Technologies | DENV-2 Den 2.2F |
| CATCTCATTGAAGTCNAGGCC | Integrated DNA Technologies | DENV-2 Den 2.2R |
| ATGAGATGYGTGGGAGTRGGAAAC | Integrated DNA Technologies | DENV-3 D3MGBEn1sFWD |
| CAC CAC DTC AAC CCA CGT AGC T | Integrated DNA Technologies | DENV-3 D3MGBEn71rREV |
| GGTGACRTTYAARGTHCCTCAT | Integrated DNA Technologies | DENV-4 D4TEn711sFWD |
| WGARTGCATRGCTCCYTCCTG | Integrated DNA Technologies | DENV-4 D4TEn786cREV |
| GARAGACCAGAGATCCTGCTGTCT | Integrated DNA Technologies | DENV-1 to 4 DV1-4_realt_fwd |
| ACCATTCCATTTTCTGGCGTT | Integrated DNA Technologies | DENV-1 to 4 DV1-4_realt_rev |
| CAG CTC CCG GAC TGA CTG | Integrated DNA Technologies | ALDOA_fwd |
| ATT CCA CGG GCT AGA GGA G | Integrated DNA Technologies | ALDOA_rev |
| GGC AAT GAC GAA GGA GGT TA | Integrated DNA Technologies | CD47_fwd |
| ATC CGG TGG TAT GGA TGA GA | Integrated DNA Technologies | CD47_rev |
| AGA AGG AAG AAA GGG CAG ACT T | Integrated DNA Technologies | CYCS_fwd |
| GGC AGT GGC CAA TTA TTA CTC a | Integrated DNA Technologies | CYCS_rev |
| AGT CAA AGA TCT CCC TGG CA | Integrated DNA Technologies | ENO1_fwd |
| TAC GTT CAC CTC GGT GTC TG | Integrated DNA Technologies | ENO1_rev |
| GCA AAT TCC ATG GCA CCG T | Integrated DNA Technologies | GAPDH_fwd |
| TCG CCC CAC TTG ATT TTG G | Integrated DNA Technologies | GAPDH_rev |
| CTT TGC TTT CCT TGG TCA GG | Integrated DNA Technologies | HPRT_fwd |
| CAA GGG CAT ATC CTA CAA CAA AC | Integrated DNA Technologies | HPRT_rev |
| CAG GTG TTC CAG AGC GAG TT | Integrated DNA Technologies | NDUFB2_fwd |
| TAC GGA AAG TGA CCC AGC AC | Integrated DNA Technologies | NDUFB2_rev |
| TAG GGC TGA GGT TTG TCC AG | Integrated DNA Technologies | SOD2_fwd |
| CAC CGA GGA GAA GTA CCA GG | Integrated DNA Technologies | SOD2_rev |
| GGC GAA GTC GAT ATA GGC AG | Integrated DNA Technologies | TPI1_fwd |
| AGT TCT TCG TTG GGG GAA AC | Integrated DNA Technologies | TPI1_rev |
| TAC GTC CCA GGA GAT GGA GA | Integrated DNA Technologies | UCP2_fwd |
| CCG TGA ACC TTA CAA AGC C | Integrated DNA Technologies | UCP2_rev |
| AAG CAG TGG TAT CAA CGC AGA GTA CAT rGrG + G | Integrated DNA Technologies | TSO |
| AAG CAG TGG TAT CAA CGC AGA GTA CT30V N | Integrated DNA Technologies | Oligo-dT30VN |
| AAG CAG TGG TAT CAA CGC AGA GT | Integrated DNA Technologies | ISPCR oligo |
| Deposited data | ||
| Human reference genome - release 22 (GRCh38.p2) | GENCODE | https://www.gencodegenes.org/human/release_22.html |
| Dataset GSE6691 | Gutiérrez et al. (2007) (Gutiérrez et al., 2007) | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6691 |
| Dataset GSE172180 | This study | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE172180 |
| Software and algorithms | ||
| FlowJo_v10 software | Tree Star | https://www.flowjo.com/ |
| scPred | Alquicira-Hernandez J et al., 2019 (Alquicira-Hernandez et al., 2019) | https://github.com/powellgenomicslab/scPred |
| Seurat | Butler et al., 2018 (Butler et al., 2018) | https://satijalab.org/seurat/ |
| Fluidigm’s SINGuLAR toolset v3.5.2 | N/A | https://www.fluidigm.com/software |
| Ingenuity Pathway Analysis (IPA) | N/A | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis |
| BASIC: BCR assembly from single cells | Canzar et al., 2017(Canzar et al., 2017) | http://ttic.uchicago.edu/∼aakhan/BASIC |
| MiXCR version 1.8.2 | Bolotin et al., 2015 (Bolotin et al., 2015) | https://github.com/milaboratory/mixcr |
| VDJtools | Shugay M et al., 2015 (Shugay et al., 2015) | https://vdjtools-doc.readthedocs.io/ |
| Gene set enrichment analysis v4.0.3 | Subramanian et al., 2005 (Subramanian et al., 2005) | https://www.gsea-msigdb.org/gsea/index.jsp |
| Connectivity Map (CMap) | Lamb et al., 2006 (Lamb et al., 2006) | https://clue.io/cmap |
| RSEM program version 1.2.19 | Li and Dewey, 2011 (Li and Dewey, 2011) | https://github.com/deweylab/RSEM |
| Harmony program | Korsunsky et al., 2019 (Korsunsky et al., 2019) | https://github.com/immunogenomics/harmony |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Katja Fink (katja.fink@alumni.ethz.ch).
Materials availability
This study did not generate new unique reagents.
Data and code availability
scRNAseq data have been deposited at GEO, accession number GSE172180. Codes used for the analysis have been published previously and are listed in the Key Resources Table.
Experimental model and subject details
Patients
Patients were recruited at Tan Tock Seng Hospital (TTSH), Singapore, following study approval from the National Healthcare Group Domain Specific Review Board (NHG DSRB Ref: 2015/0528 and DSRB B/05/013). All patients had confirmed dengue infection tested by the SD BIOLINE Dengue Duo kit (Abbott, previously Alere). The serotype of infection was determined by PCR and it remained unclear for some patients due to a Ct value close to the water control (see Table 1) . PanBio Dengue indirect IgG ELISA kit was used to assess whether the DENV infection was a primary or secondary infection. Samples used for this study were from 4 to 9 days after fever onset. Blood samples were collected in EDTA vacutainer tubes. PBMCs were isolated from peripheral blood using CPT tubes (BD) and cells were cryopreserved until use. Exclusion criteria were: (a) Positive serum test for human immunodeficiency virus (HIV), hepatitis B surface antigen (HBsAg), and/or hepatitis C antibody (b) History of receiving 1 or more doses of an investigational dengue vaccine (c) Received investigational drugs or vaccines within 2 months prior to infection (d) Hospitalization for acute illness within 3 months prior to infection (e) Known, suspected, or a history of immunocompromised (f) Use of any immunosuppressive therapy (except topical and inhaled/nebulized steroids).
Cell lines
Vero cells were purchased from ATCC and were maintained in RPMI supplemented with 10% FBS. HEK393-6E were obtained from National Research Council of Canada and were maintained in F17 medium supplemented with L-glutamine (1/50) and pluronic acid (1/111).
Method details
Single cell sorting
For Smart-seq2 sequencing: Thawed PBMCs were stained and resuspended in sorting buffer (PBS, 2% FCS, 1 mM EDTA) for sorting into 96-well PCR plates using a FACSAria (BD). Plasmablasts were sorted as CD19+, IgD−, CD27high and CD38high cells. Each well of the plates contained 2 μL of 1 mg/mL Ultrapure BSA (ThermoFisher) and 1 μL of 10 mM dNTP mix (New England Biolabs). Plates were placed on dry ice immediately after sorting and stored at −80°C until use.
For 10X genomics sequencing: Thawed PBMCs were labeled with antibodies against CD19, CD3 and a cell viability dye (LIVE/DEAD, ThermoFisher). PBMCs were stained and resuspended in sorting buffer (PBS, 2% FCS, 1 mM EDTA) for sorting of the CD3−CD19+ population into a 1.5mL Eppendorf tube containing PBS, 0.04% BSA for 10x processing.
Smart-seq2 single cell sequencing
Single cell cDNA libraries were using the Smart-Seq v2 protocol (Picelli et al., 2014) with the following modifications: 1. 1mg/ml BSA Lysis buffer (Ambion ThermoFisher Scientific, Waltham, MA, USA); 2. Use of 250 pg cDNA with 1/5 reaction of Illumina Nextera XT kit (Illumina, San Diego, CA, USA). The length distribution of the cDNA libraries was monitored using a DNA High Sensitivity Reagent Kit on the PerkinElmer Lab chip (PerkinElmer, Waltham, MA, USA). All samples were subjected to an indexed paired-end sequencing run of 2x151 cycles on an Illumina HiSeq 2500 or HiSeq 4000 system for about 1 million reads per sample.
10X genomics single cell sequencing
Cells were resuspended to a final cell concentration of 470–1600 cells/μL in PBS +0.04% BSA.
Using the 10X Genomics Chromium Controller, about 8,700-14,000 cells were encapsulated in droplets at a targeted cell recovery of 5000–8000 cells, resulting in estimated multiplet rates of 3.9–6.1%. Single cell 5′ Gene Expression libraries, V(D)J Enriched libraries were prepared using the 10X Genomics Chromium Single Cell 5′ Library & Gel Bead Kit v1, V(D)J Enrichment Kit, Human T and B cell, A Chip Kit and i7 Multiplex Kit according to the manufacturer's protocol (10X Genomics, Pleasanton, CA, USA). The libraries were subjected to an indexed paired-end sequencing run of 2x151 cycles on an Illumina HiSeq 4000 and NovaSeq 6000 (Illumina, San Diego, CA, USA) at a sequencing depth of 50,000 reads/cell for gene expression libraries and 7,000 reads/cell for V(D)J Enriched libraries.
Ig cloning, expression and purification
The mRNA of human IgG was amplified from single B cells as described previously (Appanna et al., 2016). One-step RT-PCR (OneStep RT-PCR Kit, Qiagen) was performed using forward primers in the heavy and light chain leader sequence and reverse primers in the constant region of IgG, kappa, or lambda. The PCR products were cloned into the Ptt5 mammalian expression vector (licensed from the National Research Council Biotechnology Research Institute, Montreal, QC, Canada). Alternatively, sequences were de novo synthesized and cloned into the ptt5 expression vectors (Genscript). Heavy and light chain plasmids (IgG1 format) were co-transfected into the HEK293-6E cells using 293-Transfectin (Invitrogen) in serum-free F17 medium supplemented with 20% TN1. Antibodies were purified using protein G beads (GE Healthcare).
ELISA and epitope binding assay
DENV-specific ELISAs were performed by coating high-binding 96-well plates overnight with PEG-precipitated DENV serotypes 1–4, or with recombinant E proteins produced in S2 cells as described previously (Xu et al., 2016). Plates were blocked with PBS, 0.05% Tween 20, and 3% skim milk. Supernatants from Ab-expressing HEK cells were incubated on virus-coated plates for 1 hr at room temperature before washing with PBS, 0.05% Tween 20 and detection with a secondary anti-human IgG-HRP (Sigma). Pooled plasma from dengue-immune healthy donors was used as a positive control. To determine the absolute concentration of mAbs, plates were coated with anti-human Ig Ab (Caltag). Different concentrations of a human IgG standard (Sigma) were included to generate a standard curve. TMB substrate solution (Sigma) was used for color reaction in all ELISAs. For dengue-specific ELISAs, an OD value at least 2-fold higher than the background (for cell culture supernatant from non-transfected cells) was defined as positive binding.
Epitope mapping assays were performed following the same protocol, except that 96-well plates were coated with mutated DENV-2 E proteins W101A, G106A, F108A as described previously (Xu et al., 2016). DENV-specific antibodies secreted by sorted plasmablasts were detected following the same protocol. Supernatants from plasmablasts cultured for 24hr were incubated on virus-coated plates for 1h at room temperature.
Virus neutralization assay
Monoclonal Antibodies were diluted to 20ug/ml (final concentration after addition of virus: 10ug/ml) and diluted 10-fold over 4–8 wells in a 96-well plates in duplicates. An equal volume of DENV-1 at MOI 0.01 (DENV-1-WestPac74, accession Nr. U88535.1) or DENV-2 at MOI 0.1 (TSV01, accession Nr. AF013774) was added to each well and incubated for 1hr at 37°C with 5% CO2. 100μL of the antibody-virus mixture was then added to Vero cells, seeded at a density of 20,000 cells per well the day before, and incubated for 1hr at 37°C - 5% CO2. 100μL of 10% FCS RPMI medium with 1% Penicillin/Streptomycin was added in each well and the plate was incubated for 4 days at 37°C - 5% CO2. On day 4, cells were fixed with 3.7% formalin, permeabilized with 0.1% Triton X-100, and blocked by adding 100μL of 10% FCS RPMI medium. Mouse anti-DENV E antibody (4G2) was incubated at 1μg/ml in each well for 2hr at 37°C. Cells were washed before adding secondary antibody goat-anti-mouse-HRP antibody for 1hr at 37°C. Finally, TMB substrate was added for 5 min and the reaction was stopped by adding HCl. OD was read at 450nm using an Infinite M200 Elisa reader (TECAN).
Mitochondrial function analysis
PBMCs were thawed and washed with RPMI without FBS before staining with 20uM Tetramethylrhodamine, Methyl Ester, Perchlorate (TMRM, ThermoFisher Scientific) or 200nM Mito Tracker™ Green (ThermoFisher Scientific) for 30 min at 37°C. Cells were washed, stained with LIVE/DEAD Aqua (ThermoFisher Scientific, 1/1000 in PBS 1X) for 30 min at 37°C, washed again and finally incubated for 20 min at 37°C with these antibodies: Pacific Blue-anti-CD19 (HIB19, Biolgend), PerCP-Cy5.5-anti-CD38 (HIT2, BD), AF700-anti-CD27 (O323, Biolegend), PE-Cy7-anti-IgD, PE or FITC-anti-CD47 (REA220, Miltenyi). Samples were acquired on an LSR Fortessa cytometer using FACSDiva software (BD Biosciences) and were analyzed using FlowJo_v10 software (Tree Star).
B-cell fluorospot
ELISPOT plates (Millipore MSIPN4550) were pre-wet with 50ul of 35% Ethanol for 1min, washed 3 times with PBS and coated with anti-human Ig antibody(H1700, ThermoFisher) or DENV-1 E protein or DENV-2 E protein (10ug/ml each) overnight at 4°C. Keyhole limpet hemocyanin coating (KLH, 10 μg/mL, Sigma-Aldrich) was used as negative coating control. Sorted CD47-negative plasmablasts (500, 200, 100 or 50 cells per well) or CD47-positive plasmablasts (depending on samples: between 500 and 55 cells for the samples with less cells) were directly seeded into coated ELISPOT plates and incubated for 16 hr. The plates were washed 3 times with PBS – 0.05% Tween and 3 times with PBS to remove the cells and incubated with AF488-anti-IgG (1ug/ml, Invitrogen #A11013) for 1h at room temperature. The plates were washed again, with an additional wash step with tap water, dried overnight at room temperature protected from light before reading with an IRIS ELISPOT reader (Mabtech). Ig secretion from sorted plasmablasts IgG+, IgA+, IgM+ or Igneg (based on surface expression, Sup. Figure S9) was analyzed by following the same protocol, except that the plates were coated with anti-human Ig only and 200 or 100 cells were plated. Cells from IgA+ and IgM+ fractions were detected using polyclonal AF488-anti-IgA (3 μg/mL, Goat anti-human IgA from Jackson Immunoresearch #109-005-011 labeled with Alexa Fluor 488 Antibody Labeling Kit from ThermoFisher #A20181) or PB-anti-IgM (2μg/ml, Biolegend, clone MHM-88, #314513), respectively. Cells from IgG+ and Igneg fractions were detected with AF488-anti-IgG as described above.
Quantitative real time-PCR (qRT-PCR)
Plasmablasts were sorted directly into 50ul of catch buffer (10mM Tris HCl, 1U/ul RNase inhibitor).
RNA of cells was isolated using Arcturus PicoPure RNA isolation kit (ThermoFisherScientific). RNA of cells was reverse transcribed to cDNA using SuperScript III First-Strand's oligo(dT)20 option (ThermoFisherScientific) according to the manual. qRT-PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad). Primers for targets are listed in the Key Resources Table. The concentration for each primer was 400μM. Cycling conditions of qRT-PCR were as follows: 1min at 95°C, (30s at 95°C, 1min at 55°C, 10s at 72°C)x40. For the dissociation curve: 1min at 95°C, 30s at 55°C, 30s at 95°C.
Quantification and statistical analysis
Smart-seq2 single cell analysis
Raw reads were aligned to the human reference genome GRCh38 from GENCODE release 22 (Harrow et al., 2012) using RSEM (Li and Dewey, 2011) program version 1.2.19 with default parameters. Gene expression values in transcripts per million (TPM) were calculated using the RSEM program and the human GENCODE annotation version 22. For comparison of relative gene expression between groups (violin plots), The TPM was log-normalized and then scaled using the Seurat package. Seurat version 2.0 (Butler et al., 2018) was used as the main pipeline analysis of the 5 patients from Smart-seq2 singe-cell RNA sequencing. All genes that were not detected in at least 3 cells of all our single cells were discarded, leaving 890 cells and 19,899 genes for all further analyses. Selection of highly variable genes was performed using Fluidigm's SINGuLAR toolset v3.5.2 (https://www.fluidigm.com/software). Log-normalization, Principal Component Analysis using the 400 most highly variable genes from SINGuLAR, UMAP visualization, clustering of cells and differentially expressed gene (DEG) analysis were performed using Seurat. Differential gene expression between clusters was analyzed using the likelihood-ratio test, selecting genes with an adjusted p value for the estimated fold changes <0.05. Functional analysis was generated through the use of IPA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis).
For the assembly and analysis of antibody sequences, basic version 1.0.1 (Canzar et al., 2017) and MiXCR version 1.8.2 (Bolotin et al., 2015) were used, respectively. VDJtools was used for the illustration of VJ pairing of cells in different clusters (https://vdjtools-doc.readthedocs.io/).
Gene Set Enrichment Analysis v4.0.3 (Subramanian et al., 2005) was performed to determine the enrichment score for each cluster regarding CD40L up- and down-regulated genes (genes sets from (Basso et al., 2004)). A nominal p value < 0.05 was considered statistically significant.
From the microarray dataset GSE6691 (Gutiérrez et al., 2007), comparisons between 5 healthy donor plasma cells (bone marrow) and 8 healthy donor “normal B lymphocytes” (peripheral blood) were performed using limma (Ritchie et al., 2015) pipeline. Adjusted p value <0.05 and logFC >1 were set as the threshold for DEGs. The signatures consisting of up- and downregulated genes, respectively 146 and 775 genes, were submitted to Connectivity Map (CMap) (Lamb et al., 2006) for gene set enrichment analysis of each cluster (only genes which are expressed in at least 10% of cells were used for the enrichment)
Harmony program (Korsunsky et al., 2019) was run within the Seurat workflow (default parameters) to remove the batch effect between transcriptome data from single CD47+ or CD47- cells from two secondary patients and the 890 cells from Smart-seq2 data. The top 20 normalized Harmony vectors were used as input to tSNE visualization. Fisher's Exact test with Bonferroni correction to compare the fraction of CD47 + cells contained in each cluster to the one in cluster 3.
10x genomics data analysis
scPred (Alquicira-Hernandez et al., 2019) was used to assign PBs identified based on CD38, CD27 and PRDM1 to clusters 0/1, 2 or 3 defined by smartseq.
Analysis of antibody and functional readouts
Prism software (Graphpad Prism version 8) was used for data illustration and statistical analysis. Paired or unpaired student's t test or Wilcoxon matched-pairs signed rank test was used as indicated in the figure legends.
Acknowledgment
We would like to acknowledge financial support from an A-STAR-Merck Research Laboratories (MRL) grant to K.F. This study received great help from the SIgN Immunomonitoring platform, supported by a BMRC IAF 311006 grant and BMRC transition funds #H16/99/b0/011.
Author contributions
A.R., R.A., J.L., T.L., D.S., and K.K. conducted experiments and analyzed data. M.C., M.C.L., L.T., A.T., and R.S. analyzed and visualized RNA-seq data. J.B. helped with methodology for the analysis of cellular metabolic activity. Y.-S.L. oversaw the clinical study and provided samples and clinical data. L.R., S.W.H., A.S., and J.C. oversaw research activity and planning. K.F. conceptualized the study, analyzed and visualized data, and wrote the original draft of the paper. K.F. and J.C. conceptualized and oversaw the single-cell analysis strategy. All authors reviewed the manuscript and provided feedback.
Declaration of interests
Authors declare no competing interests.
Published: May 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102482.
Supplemental information
References
- Alquicira-Hernandez J., Sathe A., Ji H.P., Nguyen Q., Powell J.E. ScPred: accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol. 2019 doi: 10.1186/s13059-019-1862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwis R.de, Williams K.L., Schmid M.A., Lai C.Y., Patel B., Smith S.A., Crowe J.E., Wang W.K., Harris E., de Silva A.M. Dengue viruses are enhanced by distinct populations of serotype cross-reactive antibodies in human immune sera. PLoS Pathog. 2014;10:e1004386. doi: 10.1371/journal.ppat.1004386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo M.A., Grifoni A., O P.H., Sidney J., Paul S., Peters B., de Silva A.D., Phillips E., Mallal S., Diehl S.A. Vol. 91. 2017. Human CD4 T cell responses to an attenuated tetravalent dengue vaccine parallel those induced by natural infection in magnitude, HLA restriction, and antigen specificity vaccines and antiviral agents crossm downloaded from; pp. 2147–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appanna R., Kg S., Xu M.H., Toh Y.X., Velumani S., Carbajo D., Lee C.Y., Zuest R., Balakrishnan T., Xu W. Plasmablasts during acute dengue infection represent a small subset of a broader virus-specific memory B cell pool. EBioMedicine. 2016;12:178–188. doi: 10.1016/j.ebiom.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpin C., Banchereau J., Liu Y.J. Memory B cells are biased towards terminal differentiation: a strategy that may prevent repertoire freezing. J. Exp. Med. 1997;186:931–940. doi: 10.1084/jem.186.6.931. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9294147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay A.N., Berg T.K.V. The interaction between signal regulatory protein alpha (SIRPα) and CD47: structure, function, and therapeutic target. Annu. Rev. Immunol. 2014;32:25–50. doi: 10.1146/annurev-immunol-032713-120142. [DOI] [PubMed] [Google Scholar]
- Basso K., Klein U., Niu H., Stolovitzky G.A., Tu Y., Califano A., Cattoretti G., Dalla-Favera R. Tracking CD40 signaling during germinal center development. Blood. 2004;104:4088–4096. doi: 10.1182/blood-2003-12-4291. [DOI] [PubMed] [Google Scholar]
- Benckert J., Schmolka N., Kreschel C., Zoller M.J., Sturm A., Wiedenmann B., Wardemann H. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J. Clin. Invest. 2011;121:1946–1955. doi: 10.1172/JCI44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi N.L., Onai N., Lanzavecchia A. A role for Toll-like receptors in acquired immunity: up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells. Blood. 2003;101:4500–4504. doi: 10.1182/blood-2002-11-3569. [DOI] [PubMed] [Google Scholar]
- Blanc P., Moro-Sibilot L., Barthly L., Jagot F., This S., De Bernard S., Buffat L., Dussurgey S., Colisson R., Hobeika E. ARTICLE Mature IgM-expressing plasma cells sense antigen and develop competence for cytokine production upon antigenic challenge. 2016. [DOI] [PMC free article] [PubMed]
- Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., Chudakov D.M. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzar S., Neu K.E., Tang Q., Wilson P.C., Khan A.A. BASIC: BCR assembly from single cells. Bioinformatics. 2017;33:425–427. doi: 10.1093/bioinformatics/btw631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M.J., Mitchell R.M., Sauteur P.M.M., Kelly D.F., Trück J. The antibody-secreting cell response to infection: kinetics and clinical applications. Front. Immunol. 2017;8:630. doi: 10.3389/fimmu.2017.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Tzarum N., Wilson I.A., Law M. VH1-69 antiviral broadly neutralizing antibodies: genetics, structures, and relevance to rational vaccine design. Curr. Opin. Virol. 2019;34:149–159. doi: 10.1016/j.coviro.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Voss J., Gamblin S.J., Codoni G., Macagno A., Jarrossay D., Vachieri S.G., Pinna D., Minola A., Vanzetta F. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- de Alwis R., Smith S.A., Olivarez N.P., Messer W.B., Huynh J.P., Wahala W.M., White L.J., Diamond M.S., Baric R.S., Crowe J.E.,J., de Silva A.M. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc. Natl. Acad. Sci. U S A. 2012;109:7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Wongwiwat W., Supasa S., Zhang X., Dai X., Rouvinski A., Jumnainsong A., Edwards C., Quyen N.T., Duangchinda T. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 2015;16:170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Niro R., Lee S.-J., Vander Heiden J.A., Elsner R.A., Trivedi N., Bannock J.M., Gupta N.T., Kleinstein S.H., Vigneault F., Gilbert T.J. Salmonella infection drives promiscuous B cell activation followed by extrafollicular affinity maturation. Immunity. 2015;43:120–131. doi: 10.1016/j.immuni.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M., Brandt S., Tinguely M., Zucol F., Bourquin J.P., Zauner L., Berger C., Bernasconi M., Speck R.F., Nadal D. Plasma cell toll-like receptor (TLR) expression differs from that of B cells, and plasma cell TLR triggering enhances immunoglobulin production. Immunology. 2009;128:573–579. doi: 10.1111/j.1365-2567.2009.03143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham N.D., Agrawal A., Waltari E., Croote D., Zanini F., Fouch M., Davidson E., Smith O., Carabajal E., Pak J.E. Broadly neutralizing human antibodies against dengue virus identified by single B cell transcriptomics. ELife. 2019;8:e52384. doi: 10.7554/elife.52384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellebedy A.H., Jackson K.J., Kissick H.T., Nakaya H.I., Davis C.W., Roskin K.M., McElroy A.K., Oshansky C.M., Elbein R., Thomas S. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat. Immunol. 2016;17:1226–1234. doi: 10.1038/ni.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G., Tan J.L., Smith S.A., de Alwis A.R., Ng T.S., Kostyuchenko V.A., Ibarra K.D., Wang J., Harris E., de Silva A. A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO Mol. Med. 2014;6:358–371. doi: 10.1002/emmm.201303404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G., Tan J.L., Smith S.A., de Alwis R., Ng T.S., Kostyuchenko V.A., Jadi R.S., Kukkaro P., de Silva A.M., Crowe J.E., Lok S.M. A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat. Commun. 2015;6:6341. doi: 10.1038/ncomms7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bates T.M., Cordeiro M.T., Nascimento E.J., Smith A.P., de Melo K.M., McBurney S.P., Evans J.D., Marques E.T.,J., Barratt-Boyes S.M. Association between magnitude of the virus-specific plasmablast response and disease severity in dengue patients. J. Immunol. 2013;190:80–87. doi: 10.4049/jimmunol.1103350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez N.C., Ocio E.M., de las Rivas J., Maiso P., Delgado M., Fermiñán E., Arcos M.J., Sánchez M.L., Hernández J.M., Miguel J.F.S. Gene expression profiling of B lymphocytes and plasma cells from Waldenström’s macroglobulinemia: comparison with expression patterns of the same cell counterparts from chronic lymphocytic leukemia, multiple myeloma and normal individuals. Leukemia. 2007;21:541–549. doi: 10.1038/sj.leu.2404520. [DOI] [PubMed] [Google Scholar]
- Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg J.S., Ridnour L.A., Dimitry J., Frazier W.A., Wink D.A., Roberts D.D. CD47 is necessary for inhibition of nitric oxide-stimulated vascular cell responses by thrombospondin-1. J. Biol. Chem. 2006;281:26069–26080. doi: 10.1074/jbc.M605040200. [DOI] [PubMed] [Google Scholar]
- Iversen R., Snir O., Stensland M., Kroll J.E., Steinsbø Ø., Korponay-Szabó I.R., Lundin K.E.A., Souza G.A.de, Sollid L.M. Strong clonal relatedness between serum and gut IgA despite different plasma cell origins. Cell Rep. 2017;20:2357–2367. doi: 10.1016/j.celrep.2017.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John A.L.S., Rathore A.P.S. Adaptive immune responses to primary and secondary dengue virus infections. Nat. Rev. Immunol. 2019;19:1. doi: 10.1038/s41577-019-0123-x. [DOI] [PubMed] [Google Scholar]
- Joo H., Coquery C., Xue Y., Gayet I., Dillon S.R., Punaro M., Zurawski G., Banchereau J., Pascual V., Oh S. Serum from patients with SLE instructs monocytes to promote IgG and IgA plasmablast differentiation. J. Exp. Med. 2012;209:1335–1348. doi: 10.1084/jem.20111644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A., Hasbold J., Fairfax K., Pridans C., Emslie D., McKenzie B.S., Lew A.M., Corcoran L.M., Hodgkin P.D., Tarlinton D.M., Nutt S.L. Initiation of plasma-cell differentiation is independent of the transcription factor blimp-1. Immunity. 2007;26:555–566. doi: 10.1016/j.immuni.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Katzelnick L.C., Gresh L., Halloran M.E., Mercado J.C., Kuan G., Gordon A., Balmaseda A., Harris E. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358:929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.-R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T., Kometani K., Ise W. Memory B cells. Nat. Rev. Immunol. 2015;15:149–159. doi: 10.1038/nri3802. [DOI] [PubMed] [Google Scholar]
- Lacza Z., Pankotai E., Busija D.W. Mitochondrial nitric oxide synthase: current concepts and controversies. Front. Biosci. (Landmark Ed.) 2009;14:4436–4443. doi: 10.2741/3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.Y., Williams K.L., Wu Y.C., Knight S., Balmaseda A., Harris E., Wang W.K. Analysis of cross-reactive antibodies recognizing the fusion loop of envelope protein and correlation with neutralizing antibody titers in Nicaraguan dengue cases. PLoS Negl. Trop. Dis. 2013;7:e2451. doi: 10.1371/journal.pntd.0002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam W.Y., Becker A.M., Kennerly K.M., Wong R., Curtis J.D., Llufrio E.M., McCommis K.S., Fahrmann J., Pizzato H.A., Nunley R.M. Mitochondrial pyruvate import promotes long-term survival of antibody-secreting plasma cells. Immunity. 2016;45:60–73. doi: 10.1016/j.immuni.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.-P., Subramanian A., Ross K.N. The connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Lavinder J.J., Wine Y., Giesecke C., Ippolito G.C., Horton A.P., Lungu O.I., Hoi K.H., DeKosky B.J., Murrin E.M., Wirth M.M. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc. Natl. Acad. Sci. U S A. 2014;111:2259–2264. doi: 10.1073/pnas.1317793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach S., Shinnakasu R., Adachi Y., Momota M., Makino-Okamura C., Yamamoto T., Ishii K.J., Fukuyama H., Takahashi Y., Kurosaki T. Requirement for memory B-cell activation in protection from heterologous influenza virus reinfection. Int. Immunol. 2019;31:771–779. doi: 10.1093/intimm/dxz049. [DOI] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Weisel F., Shlomchik M.J.B. cell receptor and CD40 signaling are rewired for synergistic induction of the c-Myc transcription factor in germinal center B cells. Immunity. 2018;48:313–326.e5. doi: 10.1016/j.immuni.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei H.E., Yoshida T., Sime W., Hiepe F., Thiele K., Manz R.A., Radbruch A., Dorner T. Blood-borne human plasma cells in steady state are derived from mucosal immune responses. Blood. 2009;113:2461–2469. doi: 10.1182/blood-2008-04-153544. blood-2008-04-153544. [DOI] [PubMed] [Google Scholar]
- Meihui X., Roland Z., Sumathy V., Farhana T., Xiu T.Y., Ramapraba A., Yu T.E., Daniela C., Paul M., Cheng-I W., Katja F. A potent neutralizing antibody with therapeutic potential against all four serotypes of dengue virus. Nat. Vaccin. 2017;2 doi: 10.1038/s41541-016-0003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesin L., Ersching J., Victora G.D. Germinal center B cell dynamics. Immunity. 2016;45:471–482. doi: 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Pinto D., Montani E., Bolli M., Garavaglia G., Sallusto F., Lanzavecchia A., Jarrossay D. A functional BCR in human IgA and IgM plasma cells. Blood. 2013;121:4110–4114. doi: 10.1182/blood-2012-09-459289. [DOI] [PubMed] [Google Scholar]
- Purtha W.E., Tedder T.F., Johnson S., Bhattacharya D., Diamond M.S. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. J. Exp. Med. 2011 doi: 10.1084/jem.20110740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvinski A., Guardado-Calvo P., Barba-Spaeth G., Duquerroy S., Vaney M.C., Kikuti C.M., Navarro Sanchez M.E., Dejnirattisai W., Wongwiwat W., Haouz A. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015;520:109–113. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- Shugay M., Bagaev D.V., Turchaninova M.A., Bolotin D.A., Britanova O.V., Putintseva E.V., Pogorelyy M.V., Nazarov V.I., Zvyagin I.V., Kirgizova V.I. VDJtools: unifying post-analysis of T cell receptor repertoires. PLOS Comput. Biol. 2015;11:e1004503. doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E., Duong V., Tawfik A., Ung S., Ly S., Casademont I., Prot M., Courtejoie N., Bleakley K., Buchy P. Increased adaptive immune responses and proper feedback regulation protect against clinical dengue. Sci. Transl Med. 2017;9 doi: 10.1126/scitranslmed.aal5088. [DOI] [PubMed] [Google Scholar]
- Spencer J., Sollid L.M. The human intestinal B-cell response. Mucosal Immunol. 2016;9:1113–1124. doi: 10.1038/mi.2016.59. [DOI] [PubMed] [Google Scholar]
- Stamper C.T., Wilson P.C. What are the primary limitations in B-cell affinity maturation, and how much affinity maturation can we drive with vaccination? Is affinity maturation a self-defeating process for eliciting broad protection? Cold Spring Harb. Perspect. Biol. 2017 doi: 10.1101/cshperspect.a028803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tas J.M.J., Mesin L., Pasqual G., Targ S., Jacobsen J.T., Mano Y.M., Chen C.S., Weill J.-C., Reynaud C.-A., Browne E.P. Visualizing antibody affinity maturation in germinal centers. Science. 2016;351:1048–1054. doi: 10.1126/science.aad3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- William J., Euler C., Christensen S., Shlomchik M.J. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 2002;297:2066–2070. doi: 10.1126/science.1073924. [DOI] [PubMed] [Google Scholar]
- Wilson P.C., Andrews S.F. Tools to therapeutically harness the human antibody response. Nat. Rev. Immunol. 2012;12:709–719. doi: 10.1038/nri3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J., Onlamoon N., Akondy R.S., Perng G.C., Polsrila K., Chandele A., Kwissa M., Pulendran B., Wilson P.C., Wittawatmongkol O. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J. Virol. 2012;86:2911–2918. doi: 10.1128/JVI.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Hadinoto V., Appanna R., Joensson K., Toh Y.X., Balakrishnan T., Ong S.H., Warter L., Leo Y.S., Wang C.I., Fink K. Plasmablasts generated during repeated dengue infection are virus glycoprotein-specific and bind to multiple virus serotypes. J. Immunol. 2012;189:5877–5885. doi: 10.4049/jimmunol.1201688. [DOI] [PubMed] [Google Scholar]
- Xu M., Zuest R., Velumani S., Tukijan F., Toh Y.X., Appanna R., Tan E.Y., Cerny D., MacAry P., Wang C.I., Fink K. A potent neutralizing antibody with therapeutic potential against all four serotypes of dengue virus. NPJ Vaccin. 2017;2:2. doi: 10.1038/s41541-016-0003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Zust R., Toh Y.X., Pfaff J.M., Kahle K.M., Davidson E., Doranz B.J., Velumani S., Tukijan F., Wang C.I., Fink K. Protective capacity of the human anamnestic antibody response during acute dengue virus infection. J. Virol. 2016;90:11122–11131. doi: 10.1128/JVI.01096-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNAseq data have been deposited at GEO, accession number GSE172180. Codes used for the analysis have been published previously and are listed in the Key Resources Table.