Figure 2.
Metabolic analyses validate use of CRC-on-Chip to model CRC progression
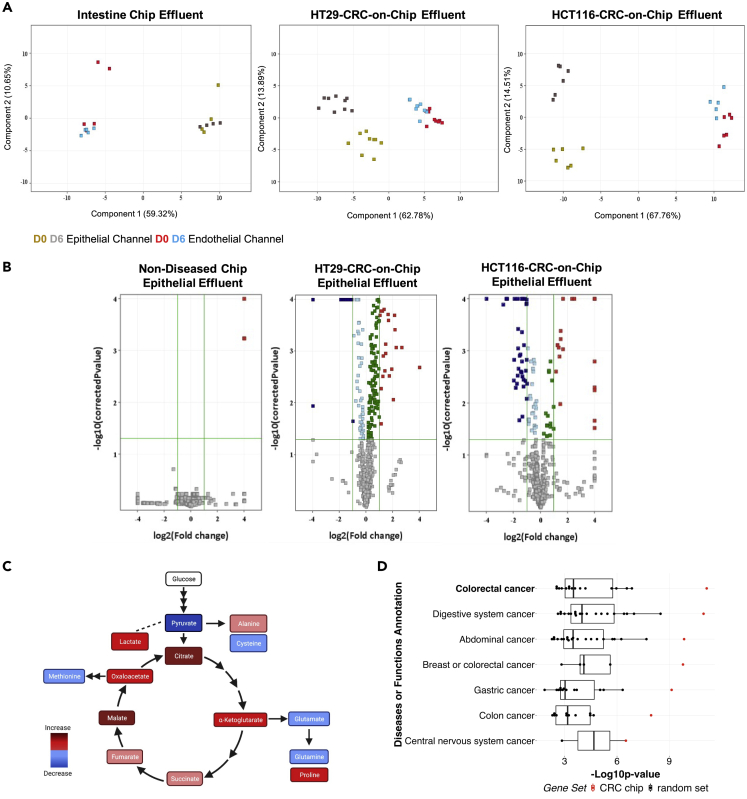
(A) Epithelial and endothelial effluent was collected from the Intestine Chip and the HT29-and HCT116-CRC-on-Chips on days 0 and 6 of the experiment and mass spectrometry-based metabolomics was performed. The principal component analysis (PCA) on the differential metabolites demonstrates the clustering of samples corresponding to the effluent compartment (epithelium or endothelium) and the different time points.
(B) Volcano plots comparing the metabolites from the top epithelial channel on day 0 and day 6 for the Intestine Chip and the HT29-and HCT116-CRC-on-Chips. Each point represents a metabolite. Analytes with p values <0.05 and fold change >2 were regarded as statistically significant (colored red and blue upregulated and downregulated, respectively).
(C) Differential metabolites between the Intestine Chip and the HCT116-CRC-on-Chip from the epithelial effluent showed altered TCA cycle and amino acid metabolism via Ingenuity Pathway Analysis (IPA).
(D) The 50 differential metabolites that matched to our internal database from the epithelial channel of the HCT116-CRC-on-Chip mapped to colorectal cancer with the highest significance (highest –Log10p value) using IPA. Each group is ranked by p value and colored based on the 50 differentially expressed metabolites from our dataset, termed “CRC Chip” (red; asterisk denotes significant outliers) or 20 permutations of 50 randomly selected metabolites, termed “random set” (black box plots).Wilcoxon signed rank test compared the CRC Chips to the random selection sets for the “colorectal cancer” disease state, p = 0.0005.