Figure 5.
The TME influences tumor cell invasion
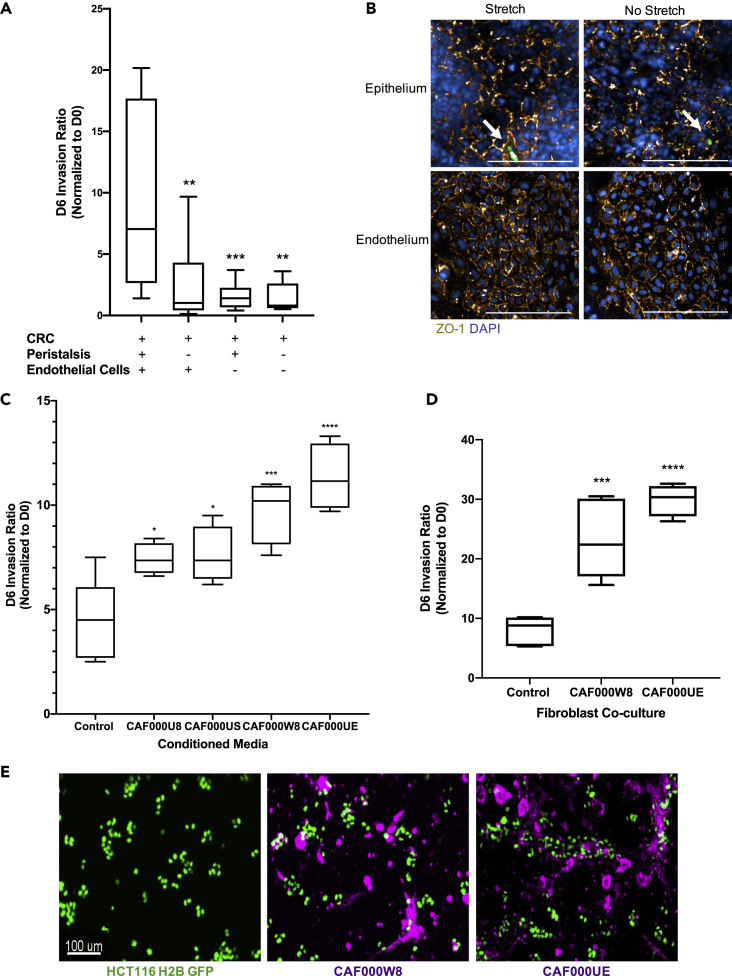
(A) CRC-on-Chips were cultured in the presence of cyclic peristalsis-like mechanical strain and HUVECs (N = 12 Chips), without cyclic strain and with HUVECs (N = 12 Chips), with cyclic strain and without HUVEC cells (N = 12 Chips) or without cyclic strain or HUVECs (N = 9 Chips). The invasion ratio of the HCT116 cells was determined via microscopy. Data are represented as boxplots and analyzed using a 1-way ANOVA with multiple comparisons; ∗∗p < 0.01, ∗∗∗p < 0.001.
(B) Representative confocal immunofluorescent images of the epithelial (top) and endothelial (bottom) channels of the CRC-on-Chips in the presence (left) or absence (right) of peristalsis stained for ZO-1 (gold) on day 6. DAPI (blue) labels the nuclei of Caco2 C2BBe1 cells in the epithelial channel and HUVECs in the endothelial channel. White arrows indicate HCT116 cells. Scale bars represent 200 μm. Images are maximum projections that span a 15 μm Z-height in the epithelial channel and a 10 μm Z-height in the endothelial channel with a 5 μm step size.
(C) Conditioned media from CAFs derived from four patients (N = 4 Chips for each patient-derived CAF) was flowed through the epithelial channel for the duration of the experiment. Differences in HCT116 cell invasion ratio quantification is depicted. Data are represented as boxplots and analyzed using 1-way ANOVA with multiple comparisons; ∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
(D) Representative CAFs derived from two patients were labeled with Cell Tracker Deep Red and seeded in the top channel prior to epithelial cell and HCT116 cell seeding (N = 4 replicates for each patient-derived CAF) and the invasion ratio was quantified on day 6. Data are represented as boxplots and analyzed using a 1-way ANOVA with multiple comparisons; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
(E) Representative images of CAF000W8 and CAF000UE on day 0 (one day after tumor cell seeding) illustrate heterocellular interactions on chip. Scale bar represents 100 μm.