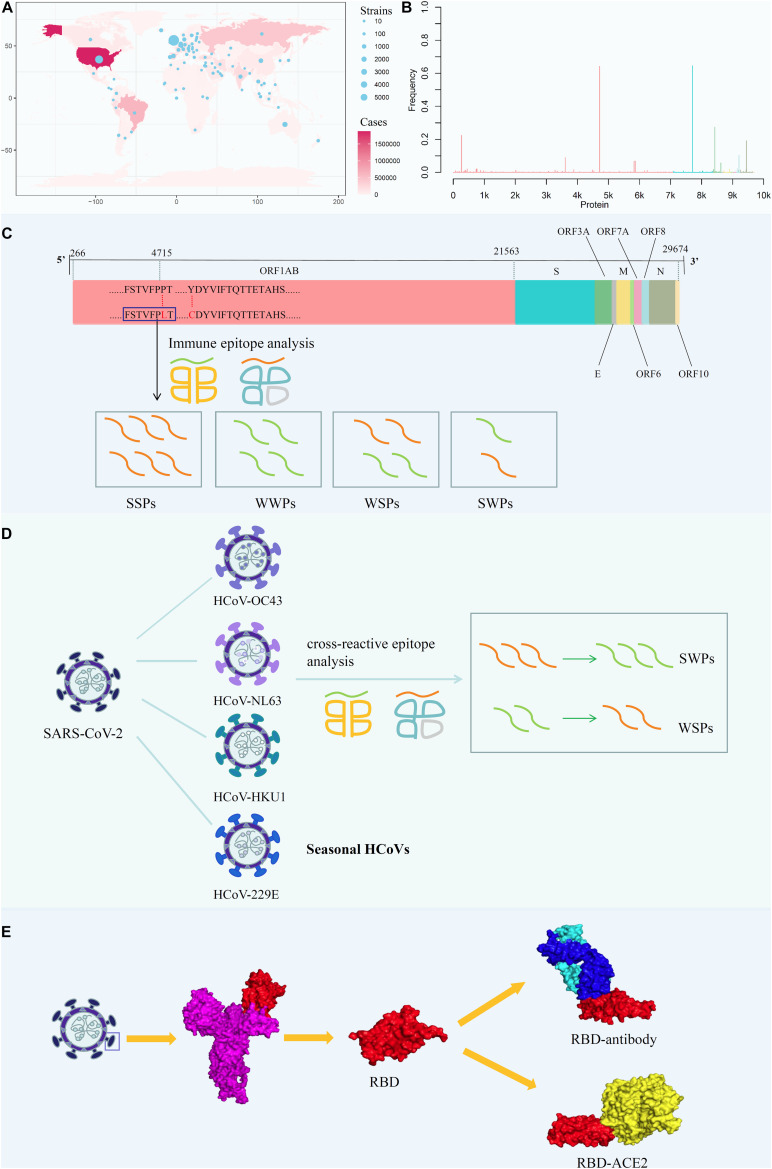
FIGURE 1.
Analysis the mutation pattern of the SARS-CoV-2 whole genome. (A) Collecting mutation strains of SARS-CoV-2 in worldwide scale. (B) Mapping the mutations on the whole genome sequence of SARS-CoV-2. (C) Deriving potential T-cell epitopes involving mutations in different structures of the whole genome sequence. According to the binding affinity predicted by IEDB standalone tools, the peptides were divided into strong binding peptide and weak binding peptide. If the mutation shifted the binding affinity of the peptides from strong to weak, it will be counted as SWPs, similarly, WSPs represents weak to strong peptides. If the binding affinity remains the same, it will be marked as SSPs and WWPs. (D) Revealing the selective pressure of cross-reactive epitopes between seasonal HCoVs and SARS-CoV-2 according to the circulating regions of viruses and the local dominant alleles. Here, by use local blast, the peptides with the same sequence between SARS-CoV-2 and other four common HCoVs were derived as cross-reactive epitopes. (E) Evaluating the binding affinity of S protein mutants against human ACE2 and binding antibody CR3022. The second subgraph is the crystal structure of SARS-CoV-2 S protein. The third subgraph is the crystal structure of the RBD region. Then, the binding complexes of RBD-ACE2 and RBD-mAb were also demonstrated.