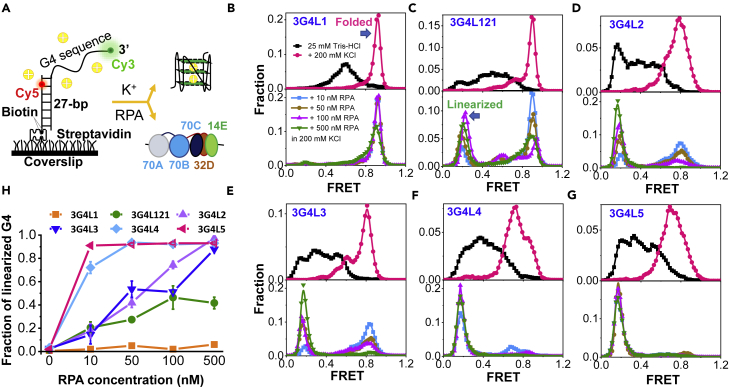
Figure 1.
RPA significantly suppresses the folding of G-rich DNA sequences into G4 structures
(A) Schematic diagram of the Cy3- and Cy5-labeled DNA harboring G4 motif. K+ will drive G4 sequence folding into G4 structure; however, on the opposite, the tight occupation by RPA (a trimer composed of 70A, 70B, and 70C domains in the 70 kD subunit; the 32 kD subunit; and the 14 kD subunit) may maintain G4 motif in the linearized state.
(B–G) The upper panels are the FRET distributions of G4 motifs in 25 mM Tris-HCl before and 4 min after the addition of 200 mM KCl with 5 mM MgCl2. The lower panels are the FRET distributions after the addition of RPA in 200 mM KCl and 5 mM MgCl2 to the G4 DNA substrates originally prepared in 25 mM Tris-HCl.
(H) The fractions of linearized G4s were obtained from the FRET peaks at ∼E0.2. The error bars were obtained from at least three repetitive experiments. Data are presented as mean ± SEM.