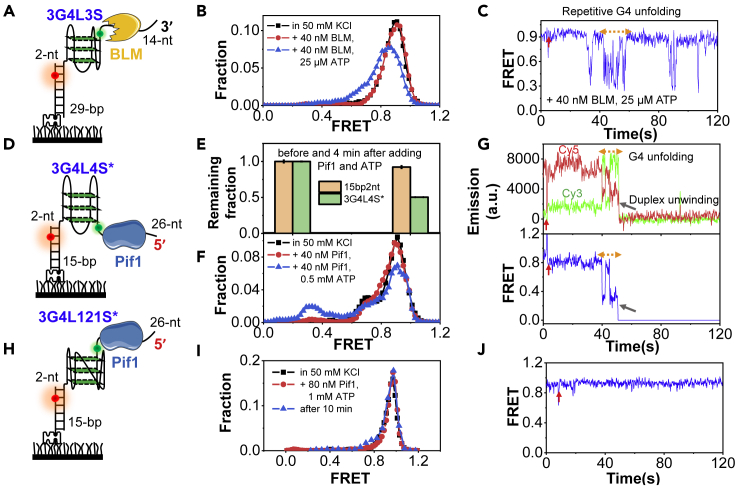
Figure 5.
The helicases-mediated unfolding of G4 DNA structures with different loop lengths
(A) The 3-layered G4 substrate with three 3-nt loops for the 3′–5′ helicase. The ssDNA tail and stem duplex are 14-nt and 29-bp, respectively.
(B) FRET distributions before and after the addition of 40 nM BLM and 25 μM ATP in 25 mM Tris-HCl, 50 mM KCl, and 5 mM MgCl2. Adding only 40 nM BLM without ATP has no detectable effect on the FRET distribution of G4.
(C) The representative trace.
(D) The 3-layered G4 substrate with three 4-nt loops for the 5′–3′ helicase. The ssDNA tail and stem duplex are 26-nt and 15-bp, respectively.
(E) The remaining fractions of 3G4L4S∗ and 15bp2nt on the coverslip surface before and 4 min after the addition of 40 nM Pif1 and 0.5 mM ATP in 25 mM Tris-HCl, 50 mM KCl, and 5 mM MgCl2. Pif1 cannot unwind the duplex directly from the 2-nt ssDNA linker. The error bars were obtained from at least three repetitive experiments. Data are presented as mean ± SEM.
(F) FRET distributions of the remaining 3G4L4S∗ molecules on coverslip before and after the addition of Pif1. Adding only 40 nM Pif1 without ATP has no obvious effect on the FRET distribution of G4.
(G) The representative traces.
(H) The 3-layered G4 substrate with three loops of 1-, 2-, and 1-nt for Pif1 helicase.
(I) FRET distributions of 3G4L121S∗ before and 4 min after the addition of 80 nM Pif1 and 1 mM ATP. Even after 10 min, there is no obvious G4 unfolding.
(J) The representative trace. No G4 unfolding can be observed.