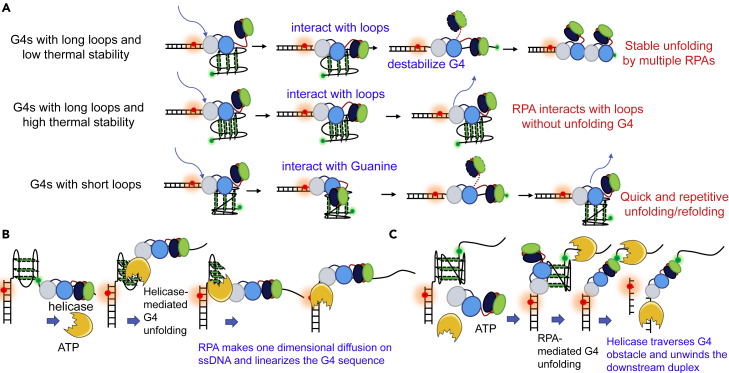
Figure 9.
The possible interacting modes of RPA with G4 structures in the absence or presence of helicases
(A) The proposed mechanisms of the interaction between RPA and different types of G4 DNA. First, for G4s with long loops (≥3-nt) and low thermal stability (Tm approximately or lower than ∼50°C), RPA may first load onto the substrate at the 4-nt linker by 70A or 70B domains. Then the trimerization core dynamically interacts with the loops, leading to the destabilization of the G4 structure. In addition, the G4 structures can be finally stably linearized by multiple RPA molecules. Second, for G4s with long loops (≥3-nt) and high thermal stability (Tm obviously higher than ∼50°C), the trimerization core predominantly interacts with the loops and the G4 structures are still intact. Third, for G4s with short loops (<3-nt), the trimerization core interacts with the Guanines in the G-tetrads, resulting in the complete or partial G4 unfolding. However, the unfolded G4s have a very strong tendency to refold back. Therefore, quick and repetitive unfolding/refolding can be observed.
(B) The proposed functions of RPA in helicases-mediated G4 unfolding. Once the G4 structure was unfolded by helicases into intermediate or ssDNA states, RPA may rapidly invade into the exposed strand by one-dimensional diffusion on the ssDNA and finally, maintain the G4 strand at the linearized state.
(C) Once the G4 was unfolded by RPA, helicase was able to traverse the G4 obstacle and further unwind the downstream duplex DNA.