Abstract
Genome editing, a revolutionary technology in molecular biology and represented by the CRISPR/Cas9 system, has become widely used in plants for characterizing gene function and crop improvement. Tomato, serving as an excellent model plant for fruit biology research and making a substantial nutritional contribution to the human diet, is one of the most important applied plants for genome editing. Using CRISPR/Cas9-mediated targeted mutagenesis, the re-evaluation of tomato genes essential for fruit ripening highlights that several aspects of fruit ripening should be reconsidered. Genome editing has also been applied in tomato breeding for improving fruit yield and quality, increasing stress resistance, accelerating the domestication of wild tomato, and recently customizing tomato cultivars for urban agriculture. In addition, genome editing is continuously innovating, and several new genome editing systems such as the recent prime editing, a breakthrough in precise genome editing, have recently been applied in plants. In this review, these advances in application of genome editing in tomato and recent development of genome editing technology are summarized, and their leaving important enlightenment to plant research and precision plant breeding is also discussed.
Keywords: CRISPR, Cas9, Solanum lycopersicum, Gene function, Precision plant breeding
Introduction
Food security is a continuous and important global issue, and increasing levels of food production are necessary due to population growth (Pradhan et al. 2015), climate change (Yu and Li 2021), and emergent global events, such as the COVID-19 pandemic (Devereux et al. 2020). With the improvement of human living standards, the increasing demand of high-quality foods with good appearance and abundant nutritional value is also put forward. Thus, technological innovation is required to meet these increasing demands of crop improvement, because conventional breeding is a rather time-consuming practice and is usually accompanied by loss of fitness and genetic diversity (Wang et al. 2019c).
Genome editing is a revolutionary technology in molecular biology and facilitates efficient, precise, and targeted modifications at genomic loci (Zhang et al. 2018). Genome editing may be the greatest innovation in plant breeding, because it allows precision plant breeding and can rapidly produce novel and transgene-free plants that are similar or identical to plants generated by conventional breeding techniques (Araki and Ishii 2015; Zhang et al. 2018). There are three major types of genome editing systems, including zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats/CRISPR-associated protein (CRISPR/Cas) systems (Zhang et al. 2018). Genome editing has been applied in different plants and is revolutionizing basic research and precision breeding in plants (Tiwari et al. 2020; Zhang et al. 2018).
As one of the most cultivated and consumed vegetable crops worldwide, tomato (Solanum lycopersicum) is rich in lycopene, vitamins, and minerals, making a substantial nutritional contribution to the human diet (Zhu et al. 2018). Tomato is also an excellent model plant for fruit biology research, and most of the knowledge about fruit development and ripening on fleshy fruit is from tomato (Pesaresi et al. 2014; Seymour et al. 2013). Genome editing has been applied in tomato since 2014 (Brooks et al. 2014; Lor et al. 2014) and has greatly facilitated the characterization of gene function and precision breeding in tomato. Noticeably, the re-evaluation of tomato genes essential for fruit ripening by using genome editing-mediated targeted mutagenesis (Gao et al. 2020; Ito et al. 2017; Wang et al. 2019b) highlights that several aspects of tomato fruit ripening should be reconsidered. Knockout mutations in several master regulators in tomato ripening generated by CRISPR/Cas9 showed weaker ripening inhibition phenotypes compared to their spontaneous mutants or RNAi plants, indicating that the regulatory network involved in fruit ripening is more complex than previously thought (Wang et al. 2020a). In addition, genome editing has been applied in tomato improvement, mainly including the improvement of fruit yield and quality, the increase of stress resistance, the domestication of wild tomato, and the customization of tomato cultivars for urban agriculture (Kwon et al. 2020; Wang et al. 2019c; Xu et al. 2019).
As a rapidly developing technology applied in basic research, plant and animal breeding, and even clinical research, genome editing is also continuously undergoing innovation (Anzalone et al. 2020), and some new genome editing systems have recently been applied in plants. In this review, these advances in application of genome editing in tomato and recent development of genome editing technology are summarized, and their leaving important enlightenment to plant research and precision plant breeding is also discussed.
Genome editing systems applied in tomato
Most genome editing systems, mainly including the three major types of genome editing systems (ZFN, TALEN, and CRISPR/Cas systems) and cytidine base editor (CBE), which belongs to be one type of DNA base editors, have been applied in tomato for basic research or precision breeding (Brooks et al. 2014; Hilioti et al. 2016; Lor et al. 2014; Shimatani et al. 2017). However, both ZFN and TALEN, which rely on protein–DNA binding to achieve sequence specificity and are generated by fusing the DNA cleavage domain of the endonuclease FokI with zinc fingers or transcriptional activator-like effectors to achieve site-specific cleavage (Xu et al. 2019), are rarely used in tomato compared to the widely used CRISPR/Cas systems.
Widely used and continuously optimized CRISPR/Cas systems
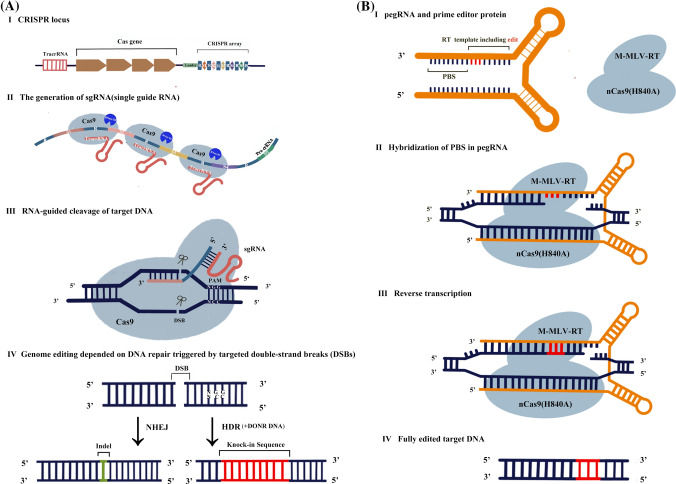
CRISPR/Cas systems are the third-generation genome editing systems, which appeared in 2012 and quickly became a superstar in genome editing tools because of their great simplicity and usability compared to with ZFN and TALEN (Zhang et al. 2018). CRISPR/Cas was originally identified as an effective acquired immune system in bacteria against virus infection and relies on RNA–DNA binding to achieve sequence specificity in genome editing (Zhang et al. 2018). CRISPR/Cas systems can be divided into five major types based on the different combinations of RNA and Cas protein, and the most studied CRISPR/Cas9 belongs to type II (Makarova et al. 2015). The CRISPR/Cas9 system is mainly composed of Cas9 protein and synthetic single-guide RNA (sgRNA) formed by the fusion of CRISPR-RNA (crRNA) and transactivation RNA (tracrRNA), and the Cas9 protein guided by sgRNA can introduce site-specific DNA double-strand breaks (DSBs), thus triggering DNA repair mechanisms (Wang et al. 2019c; Xu et al. 2019) (Fig. 1a). CRISPR/Cas9 has been applied in tomato since 2014 (Brooks et al. 2014) and has become the main genome editing tool used in tomato, from characterizing gene function to precision plant breeding (Wang et al. 2019c; Xu et al. 2019). In addition to CRISPR/Cas9, CRISPR/Cpf1(Cas12a), which belongs to type V, is a novel member of CRISPR/Cas genome editing systems (Makarova et al. 2015). The CRISPR/Cpf1 system has been gradually applied in several plant species, including rice, soybean, tobacco, maize, cotton, and, recently, tomato (Lee et al. 2019; Li et al. 2019; Tang et al. 2017, 2019; Vu et al. 2020; Wang et al. 2018) . Noticeably, the combination of CRISPR/Cpf1 and geminiviral multi-replicons significantly increased (approximately threefold) the homology-directed repair (HDR)-based genome editing efficiency in tomato compared to a Cas9-based single-replicon system (Vu et al. 2020), which demonstrates good prospects of CRISPR/Cpf1 in tomato genome editing.
Fig. 1.
Working models of conventional CRISPR/Cas9 (a) and recent prime editing (b). TracrRNA, transactivation RNA; crRNA, CRISPR-RNA; sgRNA, small-guide RNA; PAM, protospacer adjacent motif; NHEJ, non-homologous end joining; HDR, homology-directed repair; pegRNA, prime editing guide RNA; RT, reverse transcription; PBS, primer-binding site; nCas9 (H840A), catalytically impaired Cas9 (H840A) nickase (nCas9); M-MLV-RT, Moloney murine leukemia virus reverse transcriptase
Although CRISPR/Cas9 has been widely used in various organisms, numerous technological improvements (Table 1), which can improve the editing efficiency, reduce potential off-target effects, accelerate the generation of genome-edited and transgene-free plants, and confer mutations in specific cell types, tissues, or organs, have been realized to optimize the CRISPR/Cas9-mediated genome editing in plants, including tomato. Using multiplex sgRNAs instead of single sgRNA can increase the chance of obtaining large deletions in tomato mutants generated by CRISPR/Cas9 (Santillan Martinez et al. 2020). Adding an expression cassette for overexpressing an anthocyanin intensifying gene PAP1/MYB75 into the CRISPR/Cas9 construct accelerates the isolation of transgene-free tomato plants, which can be easily screened by the plant color with naked eyes (Hu et al. 2019). A spatiotemporally regulated CRISPR/Cas9 toolkit in which Cas9 expression is driven by a fruit-specific promoter (phosphoenolpyruvate carboxylase 2 gene promoter) was recently reported to confer fruit-specific gene editing in tomato (Feder et al. 2020), which will significantly accelerate fruit biology research. The potential application of important technological improvements reported in other plants (Table 1), such as the recent establishment of nanoparticle-mediated plant genome editing via a simple foliar spray of nanoparticles coated with CRISPR/Cas9 constructs (Doyle et al. 2019) and the de novo induction of gene-edited meristems for avoiding time-consuming tissue culture (Maher et al. 2020), for characterizing gene function and crop improvement remains to be investigated in tomato.
Table 1.
Summary of the important technological improvements to optimize CRISPR/Cas9-mediated genome editing in plants, including tomato
| Technological improvement | Applied in plants | References |
|---|---|---|
| Improving editing efficiency | ||
| Codon optimization of Cas9 protein by utilizing a plant codon-optimized Cas9 gene | Arabidopsis, rice and maize | Castel et al. (2019), Ma et al. (2015) and Xing et al. (2014) |
| Selection of different promoters to drive Cas9 protein | Arabidopsis and maize | Castel et al. (2019), Feng et al. (2018), Svitashev et al. (2015) and Xing et al. (2014) |
| Selection of highly efficient sgRNA | Arabidopsis and rice | Castel et al. (2019) and Liang et al. (2016) |
| Using multiplex sgRNAs instead of single sgRNA | Tomato, rice, and alfalfa | Santillan Martinez et al. (2020), Wang et al. (2016) and Wolabu et al. (2020) |
| Selection of strong terminator after Cas9 | Arabidopsis | Castel et al. (2019) |
| Optimization of T-DNA architecture | Arabidopsis | Castel et al. (2019) |
| Heat treatment | Arabidopsis | LeBlanc et al. (2018) |
| Suppressing RNA silencing | Arabidopsis | Mao et al. (2018) and Wang et al. (2019d) |
| Using chemically modified DNA as donor | Rice | Lu et al. (2020a, b) |
| Reducing potential off-target effects | ||
| Particle bombardment-mediated transient expression of CRISPR/Cas9 DNA or RNA | Wheat | Zhang et al. (2016) |
| Biolistic delivery of CRISPR/Cas9 RNPs into plant cells | Arabidopsis, tobacco, lettuce, rice, grapevine, apple, maize, wheat, petunia, Brassica oleracea and B. rapa | Liang et al. (2017), Liang et al. (2018), Malnoy et al. (2016), Murovec et al. (2018), Park and Choe (2019), Subburaj et al. (2016), Svitashev et al. (2016) and Woo et al. (2015) |
| Nanoparticle-mediated plant genome editing via a simple foliar spray of nanoparticles coating with CRISPR/Cas9 constructs | Wheat | Doyle et al. (2019) |
| Using Cas9 variants, including xCas9 and Cas9-NG | Rice | Zhong et al. (2019) |
| Using truncated sgRNA | Arabidopsis | Osakabe et al. (2016) |
| Accelerating the generation of genome-edited plants | ||
| Integration of a cassette containing the GRF4–GIF1 chimera into the CRISPR/Cas9 construct | Wheat and citrus | Debernardi et al. (2020) |
| De novo induction of gene-edited meristems for avoiding time-consuming tissue culture | Nicotiana benthamiana | Maher et al. (2020) |
| Nanoparticle-mediated plant genome editing via a simple foliar spray of nanoparticles coating with CRISPR/Cas9 constructs | Wheat | Doyle et al. (2019) |
| Adding an FT expression element into the CRISPR/Cas9 construct | Tobacco | Liu et al. (2019) |
| Accelerating the generation of transgene-free plants from edited plants | ||
| Agrobacterium-mediated transient expression of CRISPR/Cas9 in plants | Tobacco | Chen et al. (2018) |
| Biolistic delivery of CRISPR/Cas9 RNPs into plant cells | Arabidopsis, tobacco, lettuce, and rice | Luo et al. (2015) and Woo et al. (2015) |
| Drug-induced elimination of transgenes | Rice | Lu et al. (2017) |
| Programmed suicide gene-mediated self-elimination of transgenes | Rice | He et al. (2018, 2019) |
| Visible marker-assisted transgene elimination | Arabidopsis, rice, tomato, and tobacco | Chang et al. (2016), Gao et al. (2016), He et al. (2017, 2019, 2020), Liu et al. (2019) and Yu and Zhao (2019) |
| Conferring mutations in specific cell types, tissues, or organs | ||
| The CRISPR-based tissue-specific knockout system (CRISPR-TSKO) | Arabidopsis | Decaestecker et al. (2019) |
| A fiber-specific or fruit-specific promoter to drive Cas9 expression | Tomato and Arabidopsis | Feder et al. (2020) and Liang et al. (2019) |
Increasingly used cytidine base editor
DNA base editors, including cytidine base editors (CBEs) and adenine base editors (ABEs), are CRISPR/Cas9-derived tools and can produce precise single-base substitutions (C-to-T or A-to-G) in genomic DNA without the introduction of DSBs (Komor et al. 2016; Nishida et al. 2016). The CBE genome editing system, which specially confers a C–to–T base conversion in genomic DNA, consists of a cytidine deaminase fused with a Cas9 nickase (nCas9) and a uracil glycosylase inhibitor (Eid et al. 2018). CBE was firstly applied in tomato in 2017 and efficiently edited the two tomato hormone signaling genes DELLA and ETR1 with a base edition efficiency from 26.2% to 53.8% (Shimatani et al. 2017). The acetolactate synthase (ALS) gene is involved in the biosynthetic pathway for branched-chain amino acid synthesis (McCourt and Duggleby 2006), and the mutation of the Proline-186 residue in tomato ALS1 can confer chlorsulfuron resistance (Yu et al. 2010). The CBE-mediated mutation of the tomato ALS1 gene successfully created chlorsulfuron-resistant tomato plants (Veillet et al. 2019). Noticeably, the precise base edition efficiency at tomato ALS1 Pro186 codon was up to 71.4%, and 12.9% of these chlorsulfuron-resistant tomato plants were transgene-free (Veillet et al. 2019). These results highlight the feasibility of CBE for tomato genetic improvement.
Genome editing-mediated re-evaluation of tomato genes essential for fruit ripening
RNAi-mediated gene silencing or identification of the genes underlying spontaneous mutants was widely used for evaluating gene function in tomato before the emergence of genome editing systems (Wang et al. 2019b). With the establishment of different genome editing systems, especially CRISPR/Cas9, in tomato, tomato mutations can be generated rapidly, and numerous tomato genes involved in development and ripening, fruit yield and quality, and stress responses have been functionally validated using genome editing-mediated targeted mutagenesis (Xu et al. 2019). Noticeably, CRISPR/Cas9-mediated re-evaluation of tomato transcription factors and cell wall modifying enzymes in fruit ripening (Table 2) highlights that several aspects of tomato fruit ripening should be reconsidered.
Table 2.
A list of publications on the genome editing-mediated re-evaluation of tomato genes essential for fruit ripening
| Gene | Function shown by genome editing | Function shown by other tools | Con | ||||
|---|---|---|---|---|---|---|---|
| Phenotype | Tool | References | Phenotype | Tool | References | ||
| Transcription factors | |||||||
| RIN | Initiated partial ripening and showed moderate red pigmentation | CRISPR/Cas9 | Ito et al. (2017), Ito et al. (2015), Ito et al. (2020) and Li et al. (2018b, 2020d) | A near-complete inhibition of ripening phenotype | Spontaneous mutant | Vrebalov et al. (2002) | No |
| NOR | Progress of ripening was only partially affected | CRISPR/Cas9 | Gao et al. (2019b) and Wang et al. (2019b) | Progress of ripening was totally blocked | Spontaneous mutant | Kumar et al. (2018), Tigchelaar (1973) and White (2002) | No |
| CNR | Only showed 2–3 days of delayed ripening and its fruit obtained the full color finally | CRISPR/Cas9 | Gao et al. (2019b) | A colorless pericarp with strongly reduced ethylene production | Spontaneous mutant and VIGS | Manning et al. (2006) | No |
| AP2a | Initiated fruit ripening earlier, but did not fully ripen | CRISPR/Cas9 | Wang et al. (2019b) | Initiated fruit ripening earlier, but did not fully ripen | RNAi | Chung et al. (2010) and Karlova et al. (2011) | Yes |
| FUL1/FUL2 | Double mutants did not reach a red ripe color | CRISPR/Cas9 | Wang et al. (2019b) | Silencing of both genes inhibited fruit ripening | RNAi | Bemer et al. (2012) | Yes |
| Cell wall modifying enzymes | |||||||
| TBG4 | No effects on fruit firmness | CRISPR/Cas9 | Wang et al. (2019a) | Modest effects on fruit firmness | Antisense | Smith et al. (1990, 2002) | No |
| PL | Significantly enhanced fruit firmness | CRISPR/Cas9 | Wang et al. (2019a) | Significantly enhanced fruit firmness | RNAi | Uluisik et al. (2016) and Yang et al. (2017) | Yes |
| PG2a | No effects on fruit firmness | CRISPR/Cas9 | Wang et al. (2019a) | No effects on fruit firmness | Antisense | Smith et al., (1990, 2002) | Yes |
Con. consistency between function shown by genome editing and function shown by other tools, Ref. references
Revisiting the function of transcription factors in fruit ripening
Tomato is a typical climacteric fruit, and the phytohormone ethylene is critical for tomato fruit ripening (Liu et al. 2020b). There are abundant spontaneous mutants in tomato, and some of these mutants show significant ripening-deficient phenotypes (Giovannoni 2007). The identification of the genes underlying the three important ripening-deficient mutants, including ripening inhibitor (rin), nonripening (nor), and colorless nonripening (Cnr), showed that all RIN, NOR, and CNR genes are transcription factor-encoding genes (Manning et al. 2006; Tigchelaar 1973; Vrebalov et al. 2002). RIN, NOR, and CNR have long been regarded as master regulators in tomato fruit ripening via transcriptional regulation of downstream fruit ripening-related genes, such as other transcription factor-encoding genes, ethylene biosynthesis genes, and cell wall modifying enzyme-encoding genes (Seymour et al. 2013). By mainly using RNAi, a few other tomato transcription factors from different families, such as AP2a, FUL1, FUL2, TAG1, HB1, and MADS1, have been proven to also be important regulators in tomato fruit ripening (Bemer et al. 2012; Cao et al. 2020; Chen et al. 2020a, b; Chung et al. 2010; Dong et al. 2013; Itkin et al. 2009; Karlova et al. 2011; Lin et al. 2008). Thus far, the function of six transcriptional regulators, including RIN, NOR, CNR, AP2a, FUL1, and FUL2, in tomato fruit ripening has been re-evaluated using CRISPR/Cas9-mediated targeted mutagenesis (Table 2). Noticeably, CRISPR/Cas9-induced mutations in the three master regulators (RIN, NOR, and CNR) showed inconsistent ripening phenotypes as spontaneous mutants or RNAi plants (Gao et al. 2020; Ito et al. 2017; Wang et al. 2019b), which indicates that the transcriptional regulatory network involved in tomato fruit ripening should be reconsidered.
RIN has long been believed to be essential for the induction of tomato fruit ripening, because the rin spontaneous mutant showed a near-complete inhibition of ripening phenotype and did not produce red pigmentation, soften or induce an ethylene burst (Vrebalov et al. 2002). However, the CRISPR/Cas9-mediated RIN-knockout mutation still initiated partial ripening and showed moderate red pigmentation (Ito et al. 2017; Li et al. 2018b), indicating that RIN is not required for tomato ripening initiation. The specific role of RIN in tomato fruit ripening was recently investigated using the RIN-knockout mutation generated by CRISPR/Cas9, and RIN was proven to still be essential for the progression of tomato fruit ripening by inducing autocatalytic system-2 of ethylene production and subsequent full ripening (Li et al. 2020d). In addition, the NOR mutant in tomato generated by CRISPR/Cas9 also only displayed partial non-ripening phenotype (Gao et al. 2019b; Wang et al. 2019b), which is distinct from the nor spontaneous mutant (Kumar et al. 2018; White 2002). The specific role of NOR in fruit ripening was also recently investigated, mainly using the NOR-knockout mutation generated by CRISPR/Cas9, indicating that NOR still plays an important role in tomato fruit ripening by interacting with other ripening-related transcription factors and transcriptionally activating ripening-related target genes (SlACS2, SlGgpps2, and SlPL) (Gao et al. 2020). The nor spontaneous mutant is a gain-of-function mutation or a dominant-negative mutation by producing a truncated 186-amino acid protein (NOR186), which disrupts the transcriptional activation region and is unable to transcriptionally activate ripening-related target genes (Gao et al. 2020). Noticeably, the overexpression of NOR cannot completely restore the ripening phenotype of the nor spontaneous mutant (Gao et al. 2020), indicating that there are other unknown factors, resulting in the non-ripening phenotype of the nor mutant. For the CNR mutant generated by CRISPR/Cas9, it only showed two to three days of delayed ripening, and its fruit obtained the full color finally (Gao et al. 2019b), which is significantly different from the Cnr spontaneous mutant and CNR silencing plants (Manning et al. 2006). Thus, CNR is not the master regulator of fruit ripening, and its role in tomato fruit ripening remains to be investigated in the future.
Different from RIN, NOR, and CNR, three other transcription factors including AP2a, FUL1, and FUL2 have been proven to show consistent ripening phenotypes between CRISPR/Cas9-mediated mutants and RNAi plants (Wang et al. 2019b). Compared to wild-type tomato plants, both tomato AP2a mutants and AP2a-RNAi plants initiated fruit ripening earlier, but did not fully ripen (Karlova et al. 2011; Wang et al. 2019b), which confirms that AP2a is a negative regulator of tomato fruit ripening initiation. By using CRISPR/Cas9 mutagenesis, FUL1 and FUL2 were proven to show overlapping functions during fruit ripening, because the FUL1 mutant and FUL2 mutants showed no apparent differences in final overall fruit color compared with wild-type plants, but the FUL1/ FUL2 double mutants did not reach a red ripe color (Wang et al. 2019b). A similar ripening phenotype was also observed in FUL1/FUL2-RNAi plants (Bemer et al. 2012). In addition, the FUL2 mutant generated by CRISPR/Cas9 also showed a fruit development phenotype that had not been described before in FUL2-RNAi plants (Wang et al. 2019b), indicating an additional role of FUL2 in fruit development.
Overall, these evidences indicate that the three tomato spontaneous mutants (rin, nor, and Cnr) cannot be used as loss-of-function mutations, and the transcriptional regulatory network involved in tomato fruit ripening is complex and highly redundant. CRISPR/Cas9-mediated targeted mutagenesis can be used to re-evaluate other tomato transcription factors essential for fruit ripening in the future.
Revisiting the function of cell wall modifying enzymes in fruit ripening
As an important feature of fleshy fruit ripening, softening is important for fruit flavor development and overall palatability (Klee and Giovannoni 2011). Over the past 30 years, transgenic plants for silencing tomato genes encoding cell wall-related enzymes, including polygalacturonase (PG) (Cantu et al. 2008; Sheehy et al. 1988; Smith et al. 1990, 1988), pectin methylesterase (Tieman and Handa 1994; Tieman et al. 1992; Wen et al. 2013), galactanase (TBG) (Smith et al. 2002), xyloglucan endo-transglycosylase (Desilva et al. 1994), expansin (Brummell et al. 1999; Cantu et al. 2008), and pectate lyase (PL) (Uluisik et al. 2016; Yang et al. 2017), have been generated to determine which activities are involved in regulating fruit softening. Silencing of these genes usually had no or only modest effects on tomato fruit softening, but suppressing PL expression significantly inhibited tomato fruit softening (Uluisik et al. 2016; Yang et al. 2017). The role of three pectin degrading enzyme-encoding genes, including PL, PG2a, and TBG4, in tomato fruit softening was re-evaluated recently using CRISPR/Cas9-mediated targeted mutagenesis (Table 2). The PL mutant generated by CRISPR/Cas9 showed consistent ripening phenotypes to PL-RNAi plants and conferred significantly enhanced fruit firmness, which highlights a key role of PL in tomato fruit softening (Wang et al. 2019a). Previous studies showed that the silencing of PG2a and TBG4 had no and modest effects on tomato fruit firmness, respectively (Smith et al. 1990, 2002), but both PG2a and TBG4 mutations generated by CRISPR/Cas9 had no effects on tomato fruit firmness (Wang et al. 2019a). Interestingly, mutations in PG2a and TBG4 had effects on other aspects in tomato fruit ripening, including fruit color and carotenoid formation (Wang et al. 2019a), which was not described previously in their silencing plants (Smith et al. 1990, 2002).
Other aspects in fruit ripening remain to be re-evaluated
In addition to transcription factors and cell wall modifying enzymes, other aspects, mainly including ethylene biosynthesis enzymes, ethylene receptors, microRNA (miRNA), and epigenetic regulators, have also been proven to regulate fruit ripening (Chen et al. 2020a, b; Wang et al. 2020c). However, most of these aspects in tomato fruit ripening was only evaluated using the RNAi method. Thus, these aspects in fruit ripening, especially ethylene biosynthesis enzymes and receptors, should be re-evaluated using genome editing, which will provide insights into the regulatory networks of fruit ripening, especially the ethylene-mediated key regulatory hierarchy in climacteric fruit ripening.
Applications of genome editing in tomato improvement
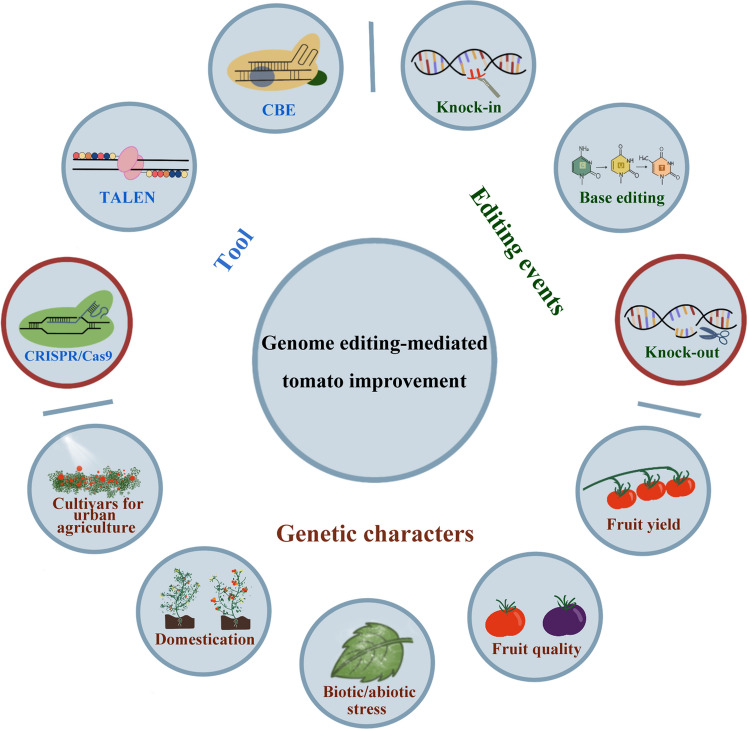
In addition to the basic research, the genetic improvement of tomato has also been greatly promoted by genome editing (Kwon et al. 2020; Wang et al. 2019c). Genome editing has been applied in tomato breeding for improving fruit yield and quality, increasing stress resistance, accelerating the domestication of wild tomato, and customizing tomato cultivars for urban agriculture by mainly using the CRISPR/Cas9 tool for creating precise knockout mutations (Fig. 2); these applications of genome editing in tomato improvement are summarized in Table 3.
Fig. 2.
A chart illustrating the applications of genome editing in tomato improvement
Table 3.
A list of publications on genome editing-mediated tomato improvement
| Targeted gene | Genome editing tool | Phenotype | References |
|---|---|---|---|
| To improve tomato fruit yield and quality | |||
| CLV3, S or SP | CRISPR/Cas9 (cis-regulatory mutations) | An increase in floral organ number or fruit size, conferring enhanced tomato fruit yield | Rodriguez-Leal et al. (2017) |
| MYB12 | CRISPR/Cas9 | Pink tomatoes | Deng et al. (2018), Yang et al. (2019) and Zhu et al. (2018) |
| CRTISO or PSY1 | CRISPR/Cas9 | Orange tomatoes and yellow tomatoes, respectively | Dahan-Meir et al. (2018) |
| SlANT1 | TALEN and CRISPR/Cas9 (targeted insertion of a strong promoter) | Purple tomatoes | Cermak et al. (2015) |
| SGR1, LCY-E, Blc, LCY-B1, and LCY-B2 | CRISPR/Cas9 | 5.1-fold increase in the lycopene content | Li et al. (2018d) |
| SlDDB1, SlDET1 and SlCYC-B | CBE | Increased carotenoid, lycopene, and β-carotene | Hunziker et al. (2020) |
| SlGAD2 and SlGAD3 | CRISPR/Cas9 | sevenfold to 15-fold increase in GABA accumulation | Nonaka et al. (2017) |
| GABA-TP1,GABA-TP2, GABA-TP3, CAT9 and SSADH | CRISPR/Cas9 | 1.34-fold to 3.50-fold in GABA accumulation | Li et al. (2018a) |
| SlIAA9 or SlAGL6 | CRISPR/Cas9 | Parthenocarpy | Ueta et al. (2017) |
| SlARF7 and SlARF5 | CRISPR/Cas9 | Parthenocarpy | Hu et al. (2018) |
| ALC | CRISPR/Cas9 | Extend long-shelf | Yu et al. (2017) |
| PL | CRISPR/Cas9 | Extend long-shelf | Uluisik et al. (2016) and Wang et al. (2019a) |
| To increase tomato resistance to stresses | |||
| SlMlo1 | CRISPR/Cas9 | Improved resistance to powdery mildew fungus Oidium neolycopersi | Nekrasov et al. (2017) |
| SLPMR4 | CRISPR/Cas9 | Improved resistance to powdery mildew fungus O. neolycopersi | Santillan Martinez et al. (2020) |
| ACET1a and ACET1b | CRISPR/Cas9 | Improved resistance to necrotrophic fungus Botrytis cinerea | Jeon et al. (2020) |
| SlDMR6-1 | CRISPR/Cas9 | Improved resistance to different oomycete or bacterial pathogens | de Toledo Thomazella et al. (2016) |
| SlJAZ2 | CRISPR/Cas9 | Improved resistance to bacterial Pto DC3000 | Ortigosa et al. (2019) |
| eIF4E1 | CRISPR/Cas9 | Improved resistance to potyvirus PepMoV | Yoon et al. (2020) |
| TYLCV genome | CRISPR/Cas9 | Improved resistance to yellow leaf curl virus TYLCV | Tashkandi et al. (2018) |
| SlLBD40 | CRISPR/Cas9 | Enhanced drought tolerance | Liu et al. (2020a) |
| ALS | CBE | Conferring resistance to sulfonylurea herbicide chlorsulfuron | Danilo et al. (2019) and Veillet et al. (2019) |
| To accelerate domestication of wild tomato | |||
| FW2.2, FAS, MULT and CycB | CRISPR/Cas9 | Threefold increase in fruit size and a tenfold increase in fruit number | Zsogon et al. (2018) |
| SP5G, SP, CLAVATA3, WUSCHEL and SlGGP1 | CRISPR/Cas9 | Conferred domesticated phenotypes yet retained parental disease resistance and salt tolerance | Li et al. (2018c) |
| To customize tomato cultivars for urban agriculture | |||
| SP, SP5G and SlER | CRISPR/Cas9 | Compactness and growth cycle of tomato plants were dramatically increased and shortened, respectively | Kwon et al. (2020) |
| HEL | CRISPR/Cas9 | Vine-like growth of tomato | Yang et al. (2020) |
Improving tomato fruit yield and quality
As one of the most important agronomic traits in crop breeding, the yield of tomato mainly depends on fruit setting rate, flowering speed, and final cell number and size of fruit, and genetic studies have identified some tomato yield-associated genes (Ariizumi et al. 2013; Zsogon et al. 2017). Inducing mutations in the promoter of the tomato signaling peptide gene CLV3, the tomato inflorescence architecture gene COMPOUND INFLORESCENCE (S), or the tomato architecture gene SELF PRUNING (SP), was generated by CRISPR/Cas9, and some of these CRISPR/Cas9 cis-regulatory mutations showed an increase in the number of floral organs or the size of fruit, conferring enhanced tomato fruit yield (Rodriguez-Leal et al. 2017). Thus, CRISPR/Cas9 drove mutagenesis of promoters will be used as a new plant precision breeding method, especially for engineering quantitative trait variation for crop improvement.
With the improvement of human living standards, greater fruit quality is increasingly needed by consumers and genome editing has also been used for the genetic improvement of tomato fruit quality (Table 3). Fruit quality is usually composed of color, size, shape, nutrients, sweetness, acidity, aroma, and also shelf life (Bai and Lindhout 2007). Although red tomatoes are the most common, tomatoes with other colors are also needed in the market for different consumers. Pink tomatoes are more popular in Asia than red tomatoes (Zhu et al. 2018), and CRISPR/Cas9-mediated targeted disruption of tomato MYB12, a master regulator of tomato flavonoid biosynthesis, in different red tomatoes successfully generated pink tomatoes (Deng et al. 2018; Yang et al. 2019; Zhu et al. 2018). CRISPR/Cas9-mediated knockout of the carotenoid isomerase (CRTISO) and phytoene synthase 1 (PSY1) genes in tomato from the carotenoid biosynthesis pathway produced orange and yellow tomatoes, respectively (Dahan-Meir et al. 2018). However, these pink, orange, and yellow tomatoes were generated at the expense of some nutrients, such as flavonoids and carotenoids. Purple tomatoes, such as the new developed tomato line “Sun Black,” are much-loved by consumers because they are rich in health-promoting anthocyanins (Blando et al. 2019). The targeted insertion of a strong promoter upstream of the endogenous anthocyanin biosynthesis gene SlANT1 in red-fruited tomatoes using CRISPR/Cas9 or TALEN produced purple tomatoes (Cermak et al. 2015), which is also an excellent case study of genome editing-mediated targeted gene insertion in plant breeding.
In addition to change visual fruit color, genome editing has also been used to improve the intrinsic quality of tomato fruit (Table 3). Tomato fruit contains abundant nutrients, such as lycopene, vitamins, and minerals, making a substantial nutritional contribution to our daily diet (Zhu et al. 2018). Multiplex editing of five genes associated with the carotenoid metabolic pathway of tomato, including stay-green 1 (SGR1), lycopene ɛ-cyclase (LCY-E), beta-lycopene cyclase (Blc), lycopene β-cyclase 1 (LCY-B1), and LCY-B2, was engineered by CRISPR/Cas9, and the multiplex-edited tomato fruit showed an approximately 5.1-fold increase in the lycopene content (Li et al. 2018d). CBE-mediated nucleotide substitutions in three other tomato genes responsible for carotenoid accumulation, including DNA Damage UV Binding protein 1 (SlDDB1), deetiolated1 (SlDET1), and Lycopene beta cyclase (SlCYC-B), also showed a significant increase in total carotenoid, lycopene, and β-carotene (Hunziker et al. 2020). Tomato fruit contains a large amount of gamma-aminobutyric acid (GABA) (Takayama and Ezura 2015), which is a non-protein amino acid and a health-promoting functional compound as an inhibitory neurotransmitter (Bachtiar et al. 2015). Multiplex-edited tomato fruit by targeting five genes involved in GABA metabolism, including three GABA-T genes (GABA-TP1, GABA-TP2, and GABA-TP3), CAT9 and SSADH, showed an increase (1.34- to 3.50-fold) in the content of GABA (Li et al. 2018a). Glutamate decarboxylase (GAD) is a key enzyme in GABA biosynthesis, and CRISPR/Cas9-induced mutations of the two tomato GAD genes SlGAD2 and SlGAD3 significantly increased GABA accumulation by sevenfold to 15-fold in tomato fruit (Nonaka et al. 2017). These results suggest that genome editing-mediated knockout or nucleotide substitutions can be used to effectively improve plant nutrients by regulating their synthesis and metabolism pathways.
Parthenocarpy is an important trait of horticultural crops because seedless fruit taste better and also have various industrial purposes (Gorguet et al. 2005; Ueta et al. 2017). Parthenocarpy is controlled by several phytohormones, especially auxin (Pandolfini 2009). CRISPR/Cas9-mediated mutations of the auxin/indole-3-acetic acid (Aux/IAA)-encoding gene SlIAA9 and two AUXIN RESPONSE FACTOR (ARF) transcription factor-encoding genes, including SlARF7 and SlARF5, produced seedless tomato fruit (Hu et al. 2018; Ueta et al. 2017), confirming the key role of auxin in parthenocarpy. Noticeably, mutations of the MADS-box gene SlAGAMOUS-LIKE 6 (SlAGL6) generated by CRISPR/Cas9 were also parthenocarpic and even showed improved yielding under heat (Klap et al. 2017). Thus, SlAGL6 is a novel and attractive gene of breeding for seedless fruit, and the specific regulation role of SlAGL6 in parthenocarpy remains to be investigated. Shelf life is also an important component of fruit quality, especially from the consumer’s point of view. CRISPR/Cas9-induced mutagenesis and replacement of tomato ALC identified from tomato spontaneous mutant alcobaca (alc) and proven to be an allele of the NOR gene (Bota et al. 2014; Garg et al. 2008), conferred a long shelf life for tomato fruit (Yu et al. 2017). The CRISPR/Cas9-induced mutation of the tomato cell wall modifying enzyme-encoding gene PL significantly improved the shelf life of tomato fruit by enhancing fruit firmness (Uluisik et al. 2016; Wang et al. 2019a). Noticeably, other fruit characteristics of AL- and CPL-edited plants were not sacrificed (Wang et al. 2019a; Yu et al. 2017), and the alc spontaneous mutant and PL-RNAi fruit even showed increased resistance to plant diseases (Bota et al. 2014; Yang et al. 2017). Thus, both the transcription factor-encoding gene ALC and the pectate lyase gene PL are valuable genetic resources for the genetic improvement of tomato.
Increasing tomato resistance to stresses
Plants are constantly subjected to numerous biotic stresses (i.e., fungi, bacteria, viruses, and nematodes as well as other plants) and abiotic stresses (i.e., drought, extreme temperatures, and chemical reagents), and there are multiple genes involved in plant responses to stresses (Ashrafi-Dehkordi et al. 2018; Bai et al. 2018). Genetic improvement of tomato for resistance to biotic and abiotic stresses is always a major objective for tomato breeders (Tieman et al. 2017). Powdery mildew, caused by the obligate biotrophic fungus Oidium neolycopersi, is a severe fungal disease for both greenhouse- and field-grown tomatoes worldwide (Jones et al. 2001), and CRISPR/Cas9-induced mutations of disease susceptibility (S) genes has been used to generate powdery mildew-resistant tomato plants (Table 3). S genes, which are required for successful pathogen infection, have received increased attention, because disabling plant S genes by genome editing is a novel breeding strategy for the genetic improvement of plant disease resistance (Pavan et al. 2010; Zaidi et al. 2018). MILDEW-RESISTANT LOCUS O (Mlo), which encodes a membrane-associated protein with seven transmembrane domains, is a conserved S gene conferring susceptibility to powdery mildew fungus in both monocots and dicots (Acevedo-Garcia et al. 2014). There are 16 Mlo genes (SlMlo1 to SlMlo16), and transgenic RNAi showed that SlMlo1 is the major contributor to powdery mildew susceptibility (Zheng et al. 2016). The CRISPR/Cas9-induced mutation of SlMlo1 significantly reduced powdery mildew susceptibility and the transgene-free powdery mildew-resistant tomato can be generated in less than 10 months (Nekrasov et al. 2017), which highlights the efficiency of genome editing in plant precision breeding. There are four Arabidopsis S genes conferring susceptibility to powdery mildew fungus, Powdery Mildew Resistant 1 (PMR1) to PMR4 (Vogel and Somerville 2000), and CRISPR/Cas9-induced mutation of SlPMR4, the closest tomato ortholog of Arabidopsis PMR4 (SlPMR4), significantly improved tomato resistance to the powdery mildew fungus O. neolycopersi (Santillan Martinez et al. 2020). Grey mold caused by the ubiquitous necrotrophic fungus Botrytis cinerea is another severe fungal disease for field- and greenhouse-grown tomatoes and can also cause serious post-harvest losses for tomato (Williamson et al. 2007). However, genome editing-mediated tomato resistance to biotic B. cinerea remains relatively unexploited. Recently, CRISPR/Cas9-induced mutations of both tomato acetylenase-encoding genes ACET1a and ACET1b showed increased resistance to B. cinerea (Jeon et al. 2020). Several candidate S genes conferring susceptibility to B. cinerea have been identified from tomato by transgenic RNAi or overexpression (Cai et al. 2020; Gonorazky et al. 2016; Sun et al. 2017), and the application of genome editing-induced mutations of these candidate S genes in breeding resistant tomato plants is promising.
Genome editing has also been used to increase tomato resistance to other different types of pathogens, including oomycetes, bacteria, and viruses (Table 3). The CRISPR/Cas9-mediated mutation of SlDMR6-1, a tomato orthologue of the Arabidopsis S gene DMR6 (Downy Mildew Resistant 6), resulted in improved resistance to different pathogens, including the oomycete pathogen Phytophthora capsici, and the bacterial pathogens Pseudomonas syringae pv. Tomato DC3000 (Pto DC3000) and Xanthomonas spp (de Toledo Thomazella et al. 2016). The CRISPR/Cas9-mediated mutation of SlJAZ2, the closest tomato ortholog of Arabidopsis JAZ (JASMONATE ZIM DOMAIN) repressor AtJAZ2, also significantly increased tomato resistance to Pto DC3000, the causal agent of tomato bacterial speck disease (Ortigosa et al. 2019). Pepper mottle virus (PepMoV) is a potyvirus that can infect tomato plants (Melzer et al. 2012), and the CRISPR/Cas9-mediated mutation of tomato eIF4E1, which encodes a eukaryotic translation initiation factor and is a recessive resistance gene against potyviruses (Wang 2015), conferred tomato resistance to PepMoV (Yoon et al. 2020). Interestingly, a stably engineered CRISPR/Cas9 system by targeting virus genes encoding coat protein (CP) or replicase (Rep) has been generated in tomato, and these tomato transgenic plants also showed significantly increased resistance to the infection of tomato yellow leaf curl virus (TYLCV) (Tashkandi et al. 2018).
Compared to genome editing-mediated tomato resistance to biotic stresses, the genetic improvement of tomato for resistance to abiotic stresses by genome editing remains relatively unexploited (Table 3). Drought is one of the most destructive abiotic stresses in plants, and many tomato cultivars are highly sensitive to drought (Zhou et al. 2017b). Recently, the CRISPR/Cas9-mediated mutation of LATERAL ORGAN BOUNDARIES DOMAIN (LBD) transcription factor-encoding gene SlLBD40 in tomato significantly enhanced tomato drought tolerance (Liu et al. 2020a), which highlights the usability of genome editing in breeding for drought tolerance in tomato. In addition, the mutation of the tomato branched-chain amino acid synthesis gene ALS1 generated by CRISPR/Cas9 and CBE showed increased resistance to the sulfonylurea herbicide chlorsulfuron (Danilo et al. 2019; Veillet et al. 2019).
Accelerating the domestication of wild tomato
Wild relatives contain some beneficial traits, such as stress tolerance, which are usually lost in conventional breeding (van de Wouw et al. 2010); thus, the de novo domestication of wild species has been proposed as an important strategy of crop improvement (Yin et al. 2017; Zsogon et al. 2017). Solanum pimpinellifolium, the putative ancestral progenitor of modern tomato varieties, is remarkably tolerant to biotic and abiotic stresses (Zuriaga et al. 2009), and genome editing has recently been used to accelerate the domestication of the wild tomato (Li et al. 2018c; Zsogon et al. 2018). Multiplex editing of four yield- and productivity-related genes, including FRUIT WEIGHT 2.2 (FW2.2), FASCIATED (FAS), MULTIFLORA (MULT), and LYCOPENE BETA CYCLASE (CycB), in the wild tomato S. pimpinellifolium was engineered by CRISPR/Cas9 (Zsogon et al. 2018). Noticeably, the multiplex-edited tomato plants showed a threefold increase in fruit size, a tenfold increase in fruit number, and a fivefold increase in fruit lycopene accumulation (Zsogon et al. 2018). In addition, CRISPR/Cas9-mediated multiplex editing of tomato genes, including flowering repressor SELF-PRUNING 5G (SP5G), SELF PRUNING (SP), the small-peptide-encoding gene CLV3 (CLAVATA3), the homeobox-encoding gene WUS (WUSCHEL), and the vitamin C-biosynthetic enzyme-encoding gene SlGGP1, also conferred domesticated phenotypes, yet retained parental disease resistance and salt tolerance in tomato plants (Li et al. 2018c). These two studies on de novo domestication of wild tomato provide a good demonstration of the domestication of wild plants using genome editing. Noticeably, genome editing has also been used for the de novo domestication of the orphan Solanaceae crop “groundcherry” (Physalis pruinosa) (Tiwari et al. 2020) and the recent allotetraploid rice (Yu et al. 2021). Based on the roadmap for de novo domestication (Fernie and Yang 2019), genome editing will significantly accelerate the de novo domestication of other wild relatives of important crops and semi- or non-cultivated plant species.
Customizing tomato cultivars for urban agriculture
The cultivation of crops in urban environments is an ideal approach for developing sustainable agriculture in the future because of the loss of arable land worldwide (Pearson et al. 2010). Multiplex editing of three tomato genes, including the classical flowering repressor gene SELF PRUNING (SP), the paralog of SP SP5G, and the regulator of tomato stem length ERECTA (SlER), was generated by CRISPR/Cas9, and the compactness and growth cycle of the triple sp, sp5g, and sler mutant were dramatically increased and shortened, respectively (Kwon et al. 2020). Noticeably, the triple-determinate mutant in cherry tomato produced its first ripe fruit at less than 40 days after transplantation (Kwon et al. 2020). Meanwhile, harvest index of the triple-determinate mutant was not significantly affected, and it grew well in indoor farm systems, including a light-emitting diode (LED) growth chamber and a self-contained, climate-controlled LED hydroponic vertical farm system (Kwon et al. 2020). Thus, these vine-like tomato plants with compact and early yielding characteristics (Kwon et al. 2020) can be developed to accommodate the plant size and space restrictions of urban agriculture. Recently, the CRISPR/Cas9-mediated mutation of the tomato HEL gene identified in a spontaneous tomato helical (hel) mutant also caused vine-like growth of tomato (Yang et al. 2020), which provides a new idea for developing urban agriculture in the future.
New genome editing systems applied in plants
Genome editing technology continuously undergoes innovation (Anzalone et al. 2020), and some new genome editing systems have recently been established and applied in plants. Herein, we mainly introduce three different types of genome editing systems, including mitochondria-targeted transcription activator-like effector nucleases (mitoTALENs) (Kazama et al. 2019), APOBEC–Cas9 fusion-induced deletion systems (AFIDs) (Wang et al. 2020b), and prime editing systems (Lin et al. 2020), because they will likely further revolutionize basic research and precision breeding in plants.
mitoTALENs for mitochondrial genome editing
In addition to nuclei, plastids (chloroplasts) and mitochondria also harbor DNA, and many important genes are present in mitochondria and plastids (Bock and Knoop 2012). Although stable transformation of plant mitochondrial genomes is still infeasible, two recent reports successfully achieved stable and heritable targeted modification of the mitochondrial genes in rice, rapeseed, and Arabidopsis by using nuclear transformation and mitoTALENs which are composed of a TALEN nuclease and a mitochondria localization signal (Arimura et al. 2020; Kazama et al. 2019). The mitochondrial gene atp6-1 encoding an ATP synthase subunit also has a copy in the nuclear genome, and mitoTALEN-mediated editing of atp6-1 resulted into mutations only in its mitochondrial copy (Arimura et al. 2020), which confirms the specificity of mitoTALEN-mediated mitochondrial genome editing. To optimize mitoTALEN-mediated mitochondrial genome editing in plants, three different promoters (i.e., CaMV35S, Ubiquitin1, and RPS5A) and two types of TALENs (i.e., conventional TALEN and compact TALEN) were compared, and the RPS5A promoter and conventional TALEN were proven to be the most efficient (Arimura et al. 2020). There are approximately 60 genes and abundant non-coding DNA in plant mitochondrial genomes (Gualberto and Newton 2017), and mitoTALEN will further unravel the role of mitochondrial genomes in plant cellular processes. In addition, engineering plastid genomes, which are also organelle genomes, has been successfully used to improve many economic and agronomic traits in different plants (Li et al. 2021). Thus, the potential application of mitoTALEN-mediated mitochondrial genome editing in crop improvement is highly anticipated.
AFIDs for precise and predictable multi-nucleotide deletions
The widely used genome editing system CRISPR/Cas9 usually results in frequent small indels, including short insertions and short deletions, which likely cannot destroy the small functional regulatory elements and domains in genomic DNA (Wang et al. 2020b). To overcome this limitation, larger deletions should be produced by genome editing systems. The recently developed APOBEC–Cas9 fusion-induced deletion systems (AFIDs), which mainly include AFID-1, AFID-2 and AFID-3, were proven to efficiently produce large (multi-nucleotide) deletions in the genomes of rice and wheat (Wang et al. 2020b). Compared to other strategies of genome editing for producing large deletions, AFIDs, especially AFID-3, have significant advantages in producing predictable deletions (Wang et al. 2020b). AFID-3 has the most additional elements including uracil DNA-glucosidase from Escherichia coli and apurinic or apyrimidinic site lyase (AP lyase) from E. coli, and approximately one-third of the deletions produced by AFID-3 are predictable (Wang et al. 2020b). These AFID systems confer precise and predictable multi-nucleotide deletions in plant genomes and will significantly accelerate the study of small functional regulatory elements and domains in genomic DNA. In addition, increasing evidences indicate that small functional regulatory elements and domains in genomic DNA, such as upstream open reading frames (uORFs) and other cis-regulatory elements, are also promising targets for crop improvement (Li et al. 2020a; Xu et al. 2017; Zhang et al. 2020; Zhou et al. 2017a), and some transcription factors function in different physiological functions by selective binding to different cis-regulatory elements (Xiao et al. 2013). Thus, AFIDs will accelerate precise plant breeding by fine-tuning of the expression of target genes, mainly at the transcriptional and translational levels.
Prime editing, a breakthrough in precise genome editing
Although genome editing is revolutionizing basic research and precision breeding in plants, most genome editing systems cannot confer precision genome editing by installing desired substitutions, insertions and deletion (Li et al. 2020c). CRISPR/Cas-mediated HDR is a previous major strategy of precision genome editing in plants (Li et al. 2020c), but HDR-mediated precision genome editing remains challenging because of the low efficiency of producing HDR and delivering DNA templates in plants (Lin et al. 2020). Prime editing, a breakthrough in precise genome editing, was recently developed in human cells (Anzalone et al. 2019) and was then quickly applied in plants by multiple research groups worldwide (Table 4) (Butt et al. 2020; Hua et al. 2020; Jiang et al. 2020b; Li et al. 2020b; Lin et al. 2020; Lu et al. 2020b; Tang et al. 2020; Xu et al. 2020a, b). Prime editing mainly consists of two parts, including the fusion of a catalytically impaired Cas9 (H840A) nickase (nCas9) and a reverse transcriptase M-MLV-RT (Moloney murine leukemia virus reverse transcriptase), and an engineered guide RNA, pegRNA (prime editing guide RNA) (Anzalone et al. 2019). pegRNA is composed of a sgRNA for targeting specific sites, a reverse transcription (RT) template for conferring predefined edits, and a primer-binding site (PBS) for allowing the 3′ end of the nicked DNA strand to hybridize to the pegRNA (Anzalone et al. 2019). As a search-and-replace genome editing tool, prime editing specifically searches the target site in genomic DNA by using the sgRNA in pegRNA and replaces it in the nCas9 (H840A)-mediated nicked DNA strand by using the reverse transcription reverse and the PBS and RT template in pegRNA (Fig. 1b) (Anzalone et al. 2019). Prime editing can produce all 12 kinds of base substitutions, which is obviously more omnipotent than DNA base editors, and can result in predefined multiple base substitutions, insertions, and deletions (Lin et al. 2020). Based on the four prime editing systems established in human cells, including PE1, PE2, PE3, and PE3b (Anzalone et al. 2019), multiple prime editing systems (Table 4), which usually undergo the optimization of codons and promoters or add other components, such as nuclear localization signals, have been established and applied in different plants. Noticeably, no off-target effects were found in prime editing in plants (Li et al. 2020b). However, these prime editing systems usually have relatively low or variable editing efficiency in plants (Table 4). The length of PBS or the RT template in pegRNA and the position of the nicking sgRNA have significant effects on the editing efficiency of prime editing systems in rice (Lin et al. 2020), and the enhancement of pegRNA expression conferred much higher prime editing efficiency (up to 53.2%) in maize (Jiang et al. 2020b). Three computational models, namely DeepPE, PE_type, and PE_position, were recently developed to predict the efficiency of pegRNA in human cells (Kim et al. 2020), and designing pegRNA using these computational models and upgrading prime editing vectors will further optimize prime editing systems.
Table 4.
Summary of the prime editing systems established in different plants
| Systems | Component description | Editing efficiencies | Applied in plants | References |
|---|---|---|---|---|
| PPE2 | Corresponding to PE2 in human cells: optimization of codons and promoters for cereal plants | 2.6%–21.8% | Wheat and rice | Lin et al. (2020) |
| PPE3 | Corresponding to PE3 in human cells: optimization of codons and promoters for cereal plants | |||
| PPE3b | Corresponding to PE3b in human cells: optimization of codons and promoters for cereal plants | |||
| PE3 | Corresponding to PE3 in human cells: optimization of codons and promoters for cereal plants, and adding two nuclear localization signals (NLSs) | 2.22%–9.38% | Rice | Li et al. (2020b) |
| PPE3-V01 | Corresponding to PE3 in human cells: optimization of codons and promoters for cereal plants, and adding two NLSs | 0.05%–1.55% | Rice | Tang et al. (2020) |
| PPE3-V02 | Corresponding to PE3 in human cells: optimization of codons and promoters for cereal plants, and adding three NLSs | |||
| PPE2-V02 | Corresponding to PE2 in human cells: optimization of codons and promoters for cereal plants, and adding three NLSs | |||
| PE-P1 | Corresponding to PE3 in human cells: optimization of codons and promoters for cereal plants, and adding two NLSs | 0%–26% | Rice | Xu et al. (2020b) |
| PE-P2 | Corresponding to PE3 in human cells: optimization of codons and promoters for cereal plants, adding two NLSs and a hygromycin phosphotransferase (Hpt), and using enhanced esgRNA | |||
| pPE2 | Corresponding to PE2 in human cells: optimization of codons and promoters for cereal plants, and adding four NLSs | 0%–31.3% | Rice | Xu et al. (2020a) |
| pPE3 | Corresponding to PE3 in human cells: optimization of codons and promoters for cereal plants, and adding four NLSs and a Hpt | |||
| pPE3b | Corresponding to PE3b in human cells: optimization of codons and promoters for cereal plants, and adding four NLSs and a Hpt | |||
| Surrogate pPE2 | Corresponding to PE2 in human cells: optimization of codons and promoters for cereal plants, and adding four NLSs and a Hpt−ATG | |||
| Sp-PE2 | Corresponding to PE2 in human cells: optimization of promoters for cereal plants and adding two NLSs | 0%–17.1% | Rice | Hua et al. (2020) |
| Sp-PE3 | Corresponding to PE3 in human cells: optimization of promoters for cereal plants and adding two NLSs | |||
| Sa-PE3 | Corresponding to PE3 in human cells: optimization of promoters for cereal plants, adding two NLSs, and using SaCas9 and Sa sgRNA | |||
| PE2 | Corresponding to PE2 in human cells: optimization of promoters for cereal plants | 0.26%–2% | Rice | Butt et al. (2020) |
| PE3 | Corresponding to PE3 in human cells: optimization of promoters for cereal plants | |||
| pCXPE01 | Corresponding to PE3 in human cells: optimization of promoters to drive pegRNA and gRNA, and adding two NLSs | 0.025%–1.66% | Tomato | Lu et al. (2020b) |
| pCXPE02 | Corresponding to PE3 in human cells: optimization of promoters to drive pegRNA and gRNA, adding two NLSs, and using a plant codon-optimized M-MLV-RT | |||
| pCXPE03 | Corresponding to PE3 in human cells: optimization of all promoters, adding two NLSs, and using a plant codon-optimized M-MLV-RT | |||
| pZ1PE3 | Corresponding to PE3 in human cells: optimization of codons and promoters for cereal plants | 6.5%–53.2% | Maize | Jiang et al. (2020b) |
| pZ1PE3b | Corresponding to PE3b in human cells: optimization of codons and promoters for cereal plants | |||
| pZ1WS | Corresponding to PE3 in human cells: optimization of codons and promoters for cereal plants, doubling the number of pegRNA expression cassettes, and using two promoter systems to drive pegRNA expression | |||
| pZ1PE3-Csy4 | On the basis of pZ1PE3, a Csy-type (CRISPR system yersinia) ribonuclease 4 (Csy4) was added | |||
| pZ1PE3b-Csy4 | On the basis of pZ1PE3b, a Csy4 was added | |||
| pZ1WS-Csy4 | On the basis of pZ1WS, a Csy4 was added |
Site-directed mutations are essential for identifying the function sites of targeted genes and for studying the biological significance of single nucleotide variants (SNVs) or single nucleotide polymorphisms (SNPs) in plants (Angaji 2009; Henikoff and Comai 2003; Schilbert et al. 2020). Compared to the time-consuming transgenic overexpression of mutated genes in the gene mutant (Jiang et al. 2020a), prime editing-mediated base substitution can conveniently create a site-directed mutation in plants. Based on this significant advantage, prime editing systems will revolutionize basic plant research by increasing the depth of research. In addition, prime editing also seems to be the best genome editing tool for achieving plant precision breeding, and prime editing-mediated introduction of elite alleles into commercialized cultivars will significantly accelerate crop improvement.
Outlook on genome editing-mediated precision breeding in tomato
Conventional breeding is a rather time-consuming practice and is usually accompanied by a loss of fitness and genetic diversity (Wang et al. 2019c), and the revolutionary genome editing technology substantially accelerates precision plant breeding (Chen et al. 2019). Tomato is one of the most important vegetable crops worldwide, and genome editing has also been applied in tomato genetic improvement (Fig. 2; Table 3). Noticeably, these applications of genome editing in the genetic improvement of tomato were performed by mainly using the CRISPR/Cas9 tool for creating precise knockout mutations (Fig. 2; Table 3). Prime editing, which is a breakthrough in precise genome editing and can install desired substitutions, insertions, and deletion, has also recently been established in tomato (Lu et al. 2020b). Thus, the precise knock-in and replacement mutations generated by other genome editing tools, such as prime editing, will further accelerate precision breeding in tomato. In addition, multiplex editing of a series of related genes usually provides better results during genome editing-mediated tomato improvement (Table 3), but CRISPR/Cas9 does not always confer successful editing in all desired tomato genes (Li et al. 2018c; Zsogon et al. 2018). The recent Cas Hybrid for Multiplexed Editing and Screening Applications (ChyMErA) genome editing system established in human cells (Gonatopoulos-Pournatzis et al. 2020) has unique advantages over multiplex editing, and thus, it is a promising approach for achieving precision tomato breeding.
Although there are numerous applications of genome editing in tomato breeding for improving several agronomic traits, mainly including yield, nutritional quality, and stress responses (Table 3), there is still a long way to go for genome editing-mediated precision breeding in tomato. The deterioration in flavor quality of modern commercial tomato varieties relative to heirloom tomato varieties is a serious problem according to consumer feedback, which can be partially attributed to the excessive concern for yield and external appearance in past tomato domestication and breeding (Tieman et al. 2017; Xu et al. 2019). The achievement of tomato genome sequencing (Sato et al. 2012), tomato pan-genome construction using genome sequences of 725 tomato phylogenetically and geographically representative accessions (Gao et al. 2019a), and recent panSV (structural variant) genome for 100 diverse tomato lines (Alonge et al. 2020) identify a vast number of genes probably controlling fruit characteristics, including fruit flavor. These abundant and valuable gene resources are of great significance for genome editing-mediated precision breeding in tomato.
Concluding remarks and future perspectives
Genome editing is revolutionizing basic research in plants because of its great simplicity and usability in creating plant mutations in genomic DNA. However, other classical research methods, including RNAi-mediated gene silencing and the identification of genes underlying spontaneous mutants, are still widely used for characterizing gene function in plants, especially in non-model plants. A series of research reports indicate that CRISPR/Cas9-induced mutations in several tomato genes essential for fruit ripening show inconsistent ripening phenotypes compared to previous spontaneous mutants or RNAi plants (Table 2), and a similar phenomenon was also reported in rice (Li et al. 2016; Yu et al. 2020). These results highlight the significance of revisiting plant genes essential for different physiological processes using genome editing. Genome editing has been successfully applied in all types of plants from monocots to dicots, and the main reason for the unsuccessful application of genome editing in some plants is the lack of a stable genetic transformation system. The latest establishment of nanoparticle-mediated plant genome editing and the de novo induction of gene-edited meristems for avoiding time-consuming tissue culture (Table 1) bring great hope for the usability of genome editing in these plants. Thus, genome editing will be the standard method for characterizing gene function in plants, especially in those plants that have successfully established genome editing systems.
Genome editing has been applied in the breeding of tomato (Fig. 2) and other important crops (Tiwari et al. 2020), and, noticeably, most of the mutants generated by genome editing are loss-of-function mutations via the knockout of targeted genes. Loss-of-function mutants have unique advantages for characterizing gene function, but most of them do not usually produce agriculturally useful phenotypes (Zhu and Qian 2020). On the contrary, gain-of-function mutations via base replacement or targeted transgene insertion show great potential for direct application in crop improvement (Zhu and Qian 2020). The DNA base editor CBE-mediated base substitution and CRISPR/Cas9-mediated targeted transgene insertion of exogenous valuable genes have been reported to increase tomato resistance to abiotic stresses and to create purple tomatoes (Table 3). In addition, the recent prime editing systems, which can confer precision genome editing by installing desired substitutions and insertions, are applied in different plants (Table 4), and using chemically modified DNA as a donor in CRISPR/Cas9 can substantially improve the efficiency of gene insertion (Table 1). Thus, broad applications of genome editing-mediated gain-of-function mutations in precision plant breeding are promising.
Although genome editing has been used to improve agronomic traits, mainly including yield, nutritional quality, and stress responses, in many different crops (Tiwari et al. 2020), several less-developed or new breeding directions need to be explored in future genome editing-mediated precision plant breeding, including (1) genome editing-mediated extension of the shelf life of horticultural products to reduce postharvest loss; (2) genome editing of plant growth genes to produce compact, synchronized ripening, and rarely dropping fruit and vegetables, which are more suitable for mechanical harvesting; (3) genome editing-mediated de novo domestication of other wild relatives of important crops and semi- or non-cultivated plant species; (4) the application of genome editing in plant molecular farming to produce high-value recombinant pharmaceutical proteins by modifying recombinant pharmaceutical proteins or directly targeting insertion of exogenous pharmaceutical protein-encoding genes; (5) the application of genome editing in plant breeding for diversification, such as conceived pungent tomatoes (Naves et al. 2019). Finally, we believe that genome editing is a key tool for achieving the long-term breeding goal of creating “smart crops” (Yu and Li 2021), which show rapid adaptation to climate change and high resistance to extreme weather conditions, as well as high yield and quality.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31972123), the Project of Chongqing Science and Technology Commission (cstc2018jcyjAX0654), the Fundamental Research Funds for the Central Universities (2020CDJQY-A074), and the Youth Top-notch Talent Support Program of Chongqing (CQYC20190525).
Author Contribution statement
YC drafted the manuscript; YC and XX wrote the manuscript; XX, XC, RL and JY performed references collection and analysis; ZL revised the manuscript.
Declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Acevedo-Garcia J, Kusch S, Panstruga R. Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 2014;204:273–281. doi: 10.1111/nph.12889. [DOI] [PubMed] [Google Scholar]
- Alonge M, Wang XG, Benoit M, Soyk S, Pereira L, Zhang L, Suresh H, Ramakrishnan S, et al. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell. 2020;182:145–161. doi: 10.1016/j.cell.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angaji SA. Single nucleotide polymorphism genotyping and its application on mapping and marker-assisted plant breeding. Afr J Biotechnol. 2009;8:908–914. [Google Scholar]
- Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki M, Ishii T. Towards social acceptance of plant breeding by genome editing. Trends Plant Sci. 2015;20:145–149. doi: 10.1016/j.tplants.2015.01.010. [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Shinozaki Y, Ezura H. Genes that influence yield in tomato. Breed Sci. 2013;63:3–13. doi: 10.1270/jsbbs.63.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura S, Ayabe H, Sugaya H, Okuno M, Tamura Y, Tsuruta Y, Watari Y, Yanase S, Yamauchi T, Itoh T, Toyoda A, Takanashi H, Tsutsumi N. Targeted gene disruption of ATP synthases 6-1 and 6-2 in the mitochondrial genome of Arabidopsis thaliana by mitoTALENs. Plant J. 2020;104:1459–1471. doi: 10.1111/tpj.15041. [DOI] [PubMed] [Google Scholar]
- Ashrafi-Dehkordi E, Alemzadeh A, Tanaka N, Razi H. Meta-analysis of transcriptomic responses to biotic and abiotic stress in tomato. PeerJ. 2018;6:e4631. doi: 10.7717/peerj.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtiar V, Near J, Johansen-Berg H, Stagg CJ. Modulation of GABA and resting state functional connectivity by transcranial direct current stimulation. Elife. 2015;4:e08789. doi: 10.7554/eLife.08789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Sunarti S, Kissoudis C, Visser RGF, van der Linden CG. The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front Plant Sci. 2018;9:801. doi: 10.3389/fpls.2018.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai YL, Lindhout P. Domestication and breeding of tomatoes: what have we gained and what can we gain in the future? Ann Bot. 2007;100:1085–1094. doi: 10.1093/aob/mcm150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M, Karlova R, Ballester AR, Tikunov YM, Bovy AG, Wolters-Arts M, Rossetto PD, Angenent GC, de Maagd RA. The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell. 2012;24:4437–4451. doi: 10.1105/tpc.112.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blando F, Berland H, Maiorano G, Durante M, Mazzucato A, Picarella ME, Nicoletti I, Gerardi C, Mita G, Andersen OM. Nutraceutical characterization of anthocyanin-rich fruits produced by "Sun Black" tomato line. Front Nut. 2019;6:133. doi: 10.3389/fnut.2019.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R, Knoop V (2012) Genomics of chloroplasts and mitochondria. Springer Science & Business Media
- Bota J, Conesa MA, Ochogavia JM, Medrano H, Francis DM, Cifre J. Characterization of a landrace collection for Tomatiga de Ramellet (Solanum lycopersicum L.) from the Balearic Islands. Genet Resour Crop Ev. 2014;61:1131–1146. doi: 10.1007/s10722-014-0096-3. [DOI] [Google Scholar]
- Brooks C, Nekrasov V, Lippman ZB, Van Eck J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014;166:1292–1297. doi: 10.1104/pp.114.247577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P. Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell. 1999;11:2203–2216. doi: 10.1105/tpc.11.11.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt H, Rao GS, Sedeek K, Aman R, Kamel R, Mahfouz M. Engineering herbicide resistance via prime editing in rice. Plant Biotechnol J. 2020;18:2370–2372. doi: 10.1111/pbi.13399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai JH, Chen T, Wang Y, Qin GZ, Tian SP. SlREM1 triggers cell death by activating an oxidative burst and other regulators. Plant Physiol. 2020;183:717–732. doi: 10.1104/pp.20.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu D, Vicente AR, Greve LC, Dewey FM, Bennett AB, Labavitch JM, Powell ALT. The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. P Natl Acad Sci USA. 2008;105:859–864. doi: 10.1073/pnas.0709813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Chen J, Yue M, Xu C, Jian W, Liu Y, Song B, Gao Y, Cheng Y, Li Z. Tomato transcriptional repressor MYB70 directly regulates ethylene-dependent fruit ripening. Plant J. 2020;104:1568–1581. doi: 10.1111/tpj.15021. [DOI] [PubMed] [Google Scholar]
- Castel B, Tomlinson L, Locci F, Yang Y, Jones JDG. Optimization of T-DNA architecture for Cas9-mediated mutagenesis in Arabidopsis. PLoS ONE. 2019;14:e0204778. doi: 10.1371/journal.pone.0204778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Baltes NJ, Cegan R, Zhang Y, Voytas DF. High-frequency, precise modification of the tomato genome. Genome Biol. 2015;16:232. doi: 10.1186/s13059-015-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang ZY, Chen ZF, Wang N, Xie G, Lu JW, Yan W, Zhou JL, Tang XY, Deng XW. Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc Natl Acad Sci USA. 2016;113:14145–14150. doi: 10.1073/pnas.1613792113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Alphonse M, Liu Q. Strategies for nonviral nanoparticle-based delivery of CRISPR/Cas9 therapeutics. WIRES Nanome Nanobi. 2020;12:e1609. doi: 10.1002/wnan.1609. [DOI] [PubMed] [Google Scholar]
- Chen K, Wang Y, Zhang R, Zhang H, Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- Chen LZ, Li W, Katin-Grazzini L, Ding J, Gu XB, Li YJ, Gu TT, Wang R, Lin XC, Deng ZN, McAvoy RJ, Gmitter FG, Deng ZA, Zhao YD, Li Y. A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic Res. 2018;5:13. doi: 10.1038/s41438-018-0023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Qin GZ, Tian SP. Regulatory network of fruit ripening: current understanding and future challenges. New Phytol. 2020;228:1219–1226. doi: 10.1111/nph.16822. [DOI] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 2010;64:936–947. doi: 10.1111/j.1365-313X.2010.04384.x. [DOI] [PubMed] [Google Scholar]
- Dahan-Meir T, Filler-Hayut S, Melamed-Bessudo C, Bocobza S, Czosnek H, Aharoni A, Levy AA. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018;95:5–16. doi: 10.1111/tpj.13932. [DOI] [PubMed] [Google Scholar]
- Danilo B, Perrot L, Mara K, Botton E, Nogue F, Mazier M. Efficient and transgene-free gene targeting using Agrobacterium-mediated delivery of the CRISPR/Cas9 system in tomato. Plant Cell Rep. 2019;38:459–462. doi: 10.1007/s00299-019-02373-6. [DOI] [PubMed] [Google Scholar]
- de Toledo Thomazella DP, Brail Q, Dahlbeck D, Staskawicz B (2016) CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. BioRxiv, 064824 [DOI] [PMC free article] [PubMed]
- Debernardi JM, Tricoli DM, Ercoli MF, Hayta S, Ronald P, Palatnik JF, Dubcovsky J. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat Biotechnol. 2020;38:1274–1279. doi: 10.1038/s41587-020-0703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaestecker W, Buono RA, Pfeiffer ML, Vangheluwe N, Jourquin J, Karimi M, Van Isterdael G, Beeckman T, Nowack MK, Jacobs TB. CRISPR-TSKO: a technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell. 2019;31:2868–2887. doi: 10.1105/tpc.19.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wang H, Sun CL, Li Q, Jiang HL, Du MM, Li CB, Li CY. Efficient generation of pink-fruited tomatoes using CRISPR/Cas9 system. J Genet Genomics. 2018;45:51–54. doi: 10.1016/j.jgg.2017.10.002. [DOI] [PubMed] [Google Scholar]
- Desilva J, Arrowsmith D, Hellyer A, Whiteman S, Robinson S. Xyloglucan endotransglycosylase and plant-growth. J Exp Bot. 1994;45:1693–1701. doi: 10.1093/jxb/45.Special_Issue.1693. [DOI] [Google Scholar]
- Devereux S, Bene C, Hoddinott J. Conceptualising COVID-19's impacts on household food security. Food Secur. 2020;12:769–772. doi: 10.1007/s12571-020-01085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong TT, Hu ZL, Deng L, Wang Y, Zhu MK, Zhang JL, Chen GP. A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol. 2013;163:1026–1036. doi: 10.1104/pp.113.224436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle C, Higginbottom K, Swift TA, Winfield M, Bellas C, Benito-Alifonso D, Fletcher T, Galan CM, Edwards K, Whitney HM (2019) A simple method for spray-on gene editing in planta. BioRxiv, 805036
- Eid A, Alshareef S, Mahfouz MM. CRISPR base editors: genome editing without double-stranded breaks. Biochemical J. 2018;475:1955–1964. doi: 10.1042/BCJ20170793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder A, Jensen S, Wang AQ, Courtney L, Middleton L, Van Eck J, Liu YS, Giovannoni JJ. Tomato fruit as a model for tissue-specific gene silencing in crop plants. Hortic Res. 2020;7:142. doi: 10.1038/s41438-020-00363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Su HD, Bai H, Wang R, Liu YL, Guo XR, Liu C, Zhang J, Yuan J, Birchler JA, Han FP. High-efficiency genome editing using a dmc1 promoter-controlled CRISPR/Cas9 system in maize. Plant Biotechnol J. 2018;16:1848–1857. doi: 10.1111/pbi.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Yang JB. De novo domestication: an alternative route toward new crops for the future. Mol Plant. 2019;12:615–631. doi: 10.1016/j.molp.2019.03.016. [DOI] [PubMed] [Google Scholar]
- Gao XH, Chen JL, Dai XH, Zhang D, Zhao YD. An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol. 2016;171:1794–1800. doi: 10.1104/pp.16.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wei W, Fan ZQ, Zhao XD, Zhang YP, Jing Y, Zhu BZ, Zhu HL, Shan W, Chen JY, Grierson D, Luo YB, Jemric T, Jiang CZ, Fu DQ. Re-evaluation of the nor mutation and the role of the NAC-NOR transcription factor in tomato fruit ripening. J Exp Bot. 2020;71:3759–3759. doi: 10.1093/jxb/eraa247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Gonda I, Sun HH, Ma QY, Bao K, Tieman DM, Burzynski-Chang EA, Fish TL, Stromberg KA, Sacks GL, Thannhauser TW, Foolad MR, Diez MJ, Blanca J, Canizares J, Xu YM, van der Knaap E, Huang SW, Klee HJ, Giovannoni JJ, Fei ZJ. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat Genet. 2019;51:1044–1051. doi: 10.1038/s41588-019-0410-2. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhu N, Zhu XF, Wu M, Jiang CZ, Grierson D, Luo YB, Shen W, Zhong SL, Fu DQ, Qu GQ. Diversity and redundancy of the ripening regulatory networks revealed by the fruitENCODE and the new CRISPR/Cas9 CNR and NOR mutants. Hortic Res. 2019;6:39. doi: 10.1038/s41438-019-0122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N, Cheema DS, Dhatt AS. Genetics of yield, quality and shelf life characteristics in tomato under normal and late planting conditions. Euphytica. 2008;159:275–288. doi: 10.1007/s10681-007-9486-3. [DOI] [Google Scholar]
- Giovannoni JJ. Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Bio. 2007;10:283–289. doi: 10.1016/j.pbi.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Gonatopoulos-Pournatzis T, Aregger M, Brown KR, Farhangmehr S, Braunschweig U, Ward HN, Ha KCH, Weiss A, Billmann M, Durbic T, Myers CL, Blencowe BJ, Moffat J. Genetic interaction mapping and exon-resolution functional genomics with a hybrid Cas9-Cas12a platform. Nat Biotechnol. 2020;38:638–648. doi: 10.1038/s41587-020-0437-z. [DOI] [PubMed] [Google Scholar]
- Gonorazky G, Guzzo MC, Abd-El-Haliem AM, Joosten MHAJ, Laxalt AM. Silencing of the tomato phosphatidylinositol-phospholipase C2 (SlPLC2) reduces plant susceptibility to Botrytis cinerea. Mol Plant Pathol. 2016;17:1354–1363. doi: 10.1111/mpp.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorguet B, van Heusden AW, Lindhout P. Parthenocarpic fruit development in tomato. Plant Biol. 2005;7:131–139. doi: 10.1055/s-2005-837494. [DOI] [PubMed] [Google Scholar]
- Gualberto JM, Newton KJ. Plant mitochondrial genomes: dynamics and mechanisms of mutation. Annu Rev Plant Biol. 2017;68:225–252. doi: 10.1146/annurev-arplant-043015-112232. [DOI] [PubMed] [Google Scholar]
- He Y, Wang R, Dai X, Zhao Y. On improving CRISPR for editing plant genes: ribozyme-mediated guide RNA production and fluorescence-based technology for isolating transgene-free mutants generated by CRISPR. Prog Mol Biol Transl. 2017;149:151–166. doi: 10.1016/bs.pmbts.2017.03.012. [DOI] [PubMed] [Google Scholar]
- He YB, Zhang T, Sun H, Zhao HD, Zhao HY. A reporter for noninvasively monitoring gene expression and plant transformation. Hortic Res. 2020;7:152. doi: 10.1038/s41438-020-00390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YB, Zhu M, Wang LH, Wu JH, Wang QY, Wang RC, Zhao YD. Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol Plant. 2018;11:1210–1213. doi: 10.1016/j.molp.2018.05.005. [DOI] [PubMed] [Google Scholar]
- He YB, Zhu M, Wang LH, Wu JH, Wang QY, Wang RC, Zhao YD. Improvements of TKC technology accelerate isolation of transgene-free CRISPR/Cas9-edited rice plants. Rice Sci. 2019;26:109–117. doi: 10.1016/j.rsci.2018.11.001. [DOI] [Google Scholar]
- Henikoff S, Comai L. Single-nucleotide mutations for plant functional genomics. Annu Rev Plant Biol. 2003;54:375–401. doi: 10.1146/annurev.arplant.54.031902.135009. [DOI] [PubMed] [Google Scholar]
- Hilioti Z, Ganopoulos I, Ajith S, Bossis I, Tsaftaris A. A novel arrangement of zinc finger nuclease system for in vivo targeted genome engineering: the tomato LEC1-LIKE4 gene case. Plant Cell Rep. 2016;35:2241–2255. doi: 10.1007/s00299-016-2031-x. [DOI] [PubMed] [Google Scholar]
- Hu JH, Israeli A, Ori N, Sun TP. The Interaction between DELLA and ARF/IAA Mediates crosstalk between gibberellin and auxin signaling to control fruit initiation in tomato. Plant Cell. 2018;30:1710–1728. doi: 10.1105/tpc.18.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N, Xian ZQ, Li N, Liu YD, Huang W, Yan F, Su DD, Chen JX, Li ZG. Rapid and user-friendly open-source CRISPR/Cas9 system for single- or multi-site editing of tomato genome. Hortic Res. 2019;6:7. doi: 10.1038/s41438-018-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua K, Jiang YW, Tao XP, Zhu JK. Precision genome engineering in rice using prime editing system. Plant Biotechnol J. 2020;18:2167–2169. doi: 10.1111/pbi.13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker J, Nishida K, Kondo A, Kishimoto S, Ariizumi T, Ezura H. Multiple gene substitution by Target-AID base-editing technology in tomato. Sci Rep. 2020;10:20471. doi: 10.1038/s41598-020-77379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 2009;60:1081–1095. doi: 10.1111/j.1365-313X.2009.04064.x. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Shima Y, Nakamura N, Kotake-Nara E, Kawasaki S, Toki S. Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nat Plants. 2017;3:866–874. doi: 10.1038/s41477-017-0041-5. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Toki S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem Biophys Res Commun. 2015;467:76–82. doi: 10.1016/j.bbrc.2015.09.117. [DOI] [PubMed] [Google Scholar]
- Ito Y, Sekiyama Y, Nakayama H, Nishizawa-Yokoi A, Endo M, Shima Y, Nakamura N, Kotake-Nara E, Kawasaki S, Hirose S, Toki S. Allelic mutations in the ripening-inhibitor locus generate extensive variation in tomato ripening. Plant Physiol. 2020;183:80–95. doi: 10.1104/pp.20.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JE, Kim JG, Fischer CR, Mehta N, Dufour-Schroif C, Wemmer K, Mudgett MB, Sattely E. A pathogen-responsive gene cluster for highly modified fatty acids in tomato. Cell. 2020;180:176–187. doi: 10.1016/j.cell.2019.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang GX, Zeng J, Li ZW, Song YB, Yan HL, He JX, Jiang YM, Duan XW. Redox regulation of the NOR transcription factor is involved in the regulation of fruit ripening in tomato. Plant Physiol. 2020;183:671–685. doi: 10.1104/pp.20.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YY, Chai YP, Lu MH, Han XL, Lin QP, Zhang Y, Zhang Q, Zhou Y, Wang XC, Gao CX, Chen QJ. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 2020;21:257. doi: 10.1186/s13059-020-02170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H, Whipps JM, Gurr SJ. The tomato powdery mildew fungus Oidium neolycopersici. Mol Plant Pathol. 2001;2:303–309. doi: 10.1046/j.1464-6722.2001.00084.x. [DOI] [PubMed] [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell. 2011;23:923–941. doi: 10.1105/tpc.110.081273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama T, Okuno M, Watari Y, Yanase S, Koizuka C, Tsuruta Y, Sugaya H, Toyoda A, Itoh T, Tsutsumi N, Toriyama K, Koizuka N, Arimura SI. Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing. Nat Plants. 2019;5:722–730. doi: 10.1038/s41477-019-0459-z. [DOI] [PubMed] [Google Scholar]
- Kim HK, Yu G, Park J, Min S, Lee S, Yoon S, Kim HH. Predicting the efficiency of prime editing guide RNAs in human cells. Nat Biotechnol. 2020;39:198–206. doi: 10.1038/s41587-020-0677-y. [DOI] [PubMed] [Google Scholar]
- Klap C, Yeshayahou E, Bolger AM, Arazi T, Gupta SK, Shabtai S, Usadel B, Salts Y, Barg R. Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol J. 2017;15:634–647. doi: 10.1111/pbi.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ. Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet. 2011;45:41–59. doi: 10.1146/annurev-genet-110410-132507. [DOI] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Tamboli V, Sharma R, Sreelakshmi Y. NAC-NOR mutations in tomato Penjar accessions attenuate multiple metabolic processes and prolong the fruit shelf life. Food Chem. 2018;259:234–244. doi: 10.1016/j.foodchem.2018.03.135. [DOI] [PubMed] [Google Scholar]
- Kwon CT, Heo J, Lemmon ZH, Capua Y, Hutton SF, Van Eck J, Park SJ, Lippman ZB. Rapid customization of Solanaceae fruit crops for urban agriculture. Nat Biotechnol. 2020;38:182–188. doi: 10.1038/s41587-019-0361-2. [DOI] [PubMed] [Google Scholar]
- LeBlanc C, Zhang F, Mendez J, Lozano Y, Chatpar K, Irish VF, Jacob Y. Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J. 2018;93:377–386. doi: 10.1111/tpj.13782. [DOI] [PubMed] [Google Scholar]
- Lee K, Zhang YX, Kleinstiver BP, Guo JA, Aryee MJ, Miller J, Malzahn A, Zarecor S, Lawrence-Dill CJ, Joung JK, Qi YP, Wang K. Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol J. 2019;17:362–372. doi: 10.1111/pbi.12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MR, Li XX, Zhou ZJ, Wu PZ, Fang MC, Pan XP, Lin QP, Luo WB, Wu GJ, Li HQ. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci. 2016;7:377. doi: 10.3389/fpls.2016.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li R, Li XD, Fu DQ, Zhu BZ, Tian HQ, Luo YB, Zhu HL. Multiplexed CRISPR/Cas9-mediated metabolic engineering of gamma-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol J. 2018;16:415–427. doi: 10.1111/pbi.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xu HJL, Ju Z, Cao DY, Zhu HL, Fu DQ, Grierson D, Qin GZ, Luo YB, Zhu BZ. The RIN-MC fusion of MADS-box transcription factors has transcriptional activity and modulates expression of many ripening genes. Plant Physiol. 2018;176:891–909. doi: 10.1104/pp.17.01449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TD, Yang XP, Yu Y, Si XM, Zhai XW, Zhang HW, Dong WX, Gao CX, Xu C. Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol. 2018;36:1160–1163. doi: 10.1038/nbt.4273. [DOI] [PubMed] [Google Scholar]
- Li XD, Wang YN, Chen S, Tian HQ, Fu DQ, Zhu BZ, Luo YB, Zhu HL. Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front Plant Sci. 2018;9:559. doi: 10.3389/fpls.2018.00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Rui HP, Li YJ, Wang QQ, Alariqi M, Qin L, Sun L, Ding X, Wang FQ, Zou JW, Wang YQ, Yuan DJ, Zhang XL, Jin SX. Robust CRISPR/Cpf1 (Cas12a)-mediated genome editing in allotetraploid cotton (Gossypium hirsutum) Plant Biotechnol J. 2019;17:862–1864. doi: 10.1111/pbi.13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Li W, Zhou ZH, Chen H, Xie CH, Lin YJ. A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotechnol J. 2020;18:313–315. doi: 10.1111/pbi.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HY, Li JY, Chen JL, Yan L, Xia LQ. Precise Modifications of both exogenous and endogenous genes in rice by prime editing. Mol Plant. 2020;13:671–674. doi: 10.1016/j.molp.2020.03.011. [DOI] [PubMed] [Google Scholar]
- Li JY, Li HY, Chen JL, Yan L, Xia LQ. Toward precision genome editing in crop plants. Mol Plant. 2020;13:811–813. doi: 10.1016/j.molp.2020.04.008. [DOI] [PubMed] [Google Scholar]
- Li S, Zhu B, Pirrello J, Xu C, Zhang B, Bouzayen M, Chen K, Grierson D. Roles of RIN and ethylene in tomato fruit ripening and ripening-associated traits. New Phytol. 2020;226:460–475. doi: 10.1111/nph.16362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chang L, Zhang J (2021) Advancing organelle genome transformation and editing for crop improvement. Plant Commun 100141. 10.1016/j.xplc.2021.100141. [DOI] [PMC free article] [PubMed]
- Liang G, Zhang HM, Lou DJ, Yu DQ. Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci Rep. 2016;6:21451. doi: 10.1038/srep21451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Eudes A, Yogiswara S, Jing BB, Benites VT, Yamanaka R, Cheng-Yue C, Baidoo EE, Mortimer JC, Scheller HV, Loque D. A screening method to identify efficient sgRNAs in Arabidopsis, used in conjunction with cell-specific lignin reduction. Biotechnol Biofuels. 2019;12:130. doi: 10.1186/s13068-019-1467-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Chen KL, Li TD, Zhang Y, Wang YP, Zhao Q, Liu JX, Zhang HW, Liu CM, Ran YD, Gao CX. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun. 2017;8:14261. doi: 10.1038/ncomms14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZF, Hong YG, Yin MG, Li CY, Zhang K, Grierson D. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 2008;55:301–310. doi: 10.1111/j.1365-313X.2008.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin QP, Zong Y, Xue CX, Wang SX, Jin S, Zhu ZX, Wang YP, Anzalone AV, Raguram A, Doman JL, Liu DVR, Gao CX. Prime genome editing in rice and wheat. Nat Biotechnol. 2020;38:582–585. doi: 10.1038/s41587-020-0455-x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zeng J, Yuan C, Guo Y, Yu H, Li Y, Huang C. Cas9-PF, an early flowering and visual selection marker system, enhances the frequency of editing event occurrence and expedites the isolation of genome-edited and transgene-free plants. Plant Biotechnol J. 2019;17:1191–1193. doi: 10.1111/pbi.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang J, Xu J, Li Y, Guo L, Wang Z, Zhang X, Zhao B, Guo YD, Zhang N. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 2020;301:110683. doi: 10.1016/j.plantsci.2020.110683. [DOI] [PubMed] [Google Scholar]
- Liu YD, Tang MF, Liu MC, Su DD, Chen J, Gao YS, Bouzayen M, Li ZG. The molecular regulation of ethylene in fruit ripening. Small Methods. 2020;4:1900485. doi: 10.1002/smtd.201900485. [DOI] [Google Scholar]
- Lor VS, Starker CG, Voytas DF, Weiss D, Olszewski NE. Targeted mutagenesis of the tomato PROCERA gene using transcription activator-Like effector nucleases. Plant Physiol. 2014;166:1288–1291. doi: 10.1104/pp.114.247593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu HP, Liu SM, Xu SL, Chen WY, Zhou X, Tan YY, Huang JZ, Shu QY. CRISPR-S: an active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants. Plant Biotechnol J. 2017;15:1371–1373. doi: 10.1111/pbi.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Tian Y, Shen R, Yao Q, Wang M, Chen M, Dong J, Zhang T, Li F, Lei M, Zhu JK. Targeted, efficient sequence insertion and replacement in rice. Nat Biotechnol. 2020;38:1402–1407. doi: 10.1038/s41587-020-0581-5. [DOI] [PubMed] [Google Scholar]
- Lu YM, Tian YF, Shen RD, Yao Q, Zhong DT, Zhang XN, Zhu JK. Precise genome modification in tomato using an improved prime editing system. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Li J, Stoddard TJ, Baltes NJ, Demorest ZL, Clasen BM, Coffman A, Retterath A, Mathis L, Voytas DF, Zhang F. Non-transgenic plant genome editing using purified sequence-specific nucleases. Mol Plant. 2015;8:1425–1427. doi: 10.1016/j.molp.2015.05.012. [DOI] [PubMed] [Google Scholar]
- Ma XL, Zhang QY, Zhu QL, Liu W, Chen Y, Qiu R, Wang B, Yang ZF, Li HY, Lin YR, Xie YY, Shen RX, Chen SF, Wang Z, Chen YL, Guo JX, Chen LT, Zhao XC, Dong ZC, Liu YG. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot Plants. Mol Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Maher MF, Nasti RA, Vollbrecht M, Starker CG, Clark MD, Voytas DF. Plant gene editing through de novo induction of meristems. Nat Biotechnol. 2020;38:84–89. doi: 10.1038/s41587-019-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoy M, Viola R, Jung MH, Koo OJ, Kim S, Kim JS, Velasco R, Kanchiswamy CN. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 pibonucleoproteins. Front Plant Sci. 2016;7:1904. doi: 10.3389/fpls.2016.01904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- Mao Y, Yang X, Zhou Y, Zhang Z, Botella JR, Zhu JK. Manipulating plant RNA-silencing pathways to improve the gene editing efficiency of CRISPR/Cas9 systems. Genome Biol. 2018;19:149. doi: 10.1186/s13059-018-1529-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt JA, Duggleby RG. Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids. Amino Acids. 2006;31:173–210. doi: 10.1007/s00726-005-0297-3. [DOI] [PubMed] [Google Scholar]
- Melzer MJ, Sugano JS, Cabanas D, Dey KK, Kandouh B, Mauro D, Rushanaedy I, Srivastava S, Watanabe S, Borth WB, Tripathi S, Matsumoto T, Keith L, Gonsalves D, Hu JS. First report of pepper mottle virus infecting tomato in Hawaii. Plant Dis. 2012;96:917–917. doi: 10.1094/PDIS-02-12-0147-PDN. [DOI] [PubMed] [Google Scholar]
- Murovec J, Gucek K, Bohanec B, Avbelj M, Jerala R. DNA-free genome editing of Brassica oleracea and B. rapa protoplasts using CRISPR-Cas9 ribonucleoprotein complexes. Front Plant Sci. 2018;9:1594. doi: 10.3389/fpls.2018.01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naves ER, de Avila SL, Sulpice R, Araujo WL, Nunes-Nesi A, Peres LEP, Zsogon A. Capsaicinoids: pungency beyond capsicum. Trends Plant Sci. 2019;24:109–120. doi: 10.1016/j.tplants.2018.11.001. [DOI] [PubMed] [Google Scholar]
- Nekrasov V, Wang CM, Win J, Lanz C, Weigel D, Kamoun S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep. 2017;7:482. doi: 10.1038/s41598-017-00578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353:aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Arai C, Takayama M, Matsukura C, Ezura H. Efficient increase of gamma-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci Rep. 2017;7:7057. doi: 10.1038/s41598-017-06400-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortigosa A, Gimenez-Ibanez S, Leonhardt N, Solano R. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol J. 2019;17:665–673. doi: 10.1111/pbi.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Watanabe T, Sugano SS, Ueta R, Ishihara R, Shinozaki K, Osakabe K. Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci Rep. 2016;6:26685. doi: 10.1038/srep26685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfini T. Seedless fruit production by hormonal regulation of fruit set. Nutrients. 2009;1:168–177. doi: 10.3390/nu1020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Choe S. DNA-free genome editing with preassembled CRISPR/Cas9 ribonucleoproteins in plants. Transgenic Res. 2019;28:61–64. doi: 10.1007/s11248-019-00136-3. [DOI] [PubMed] [Google Scholar]
- Pavan S, Jacobsen E, Visser RGF, Bai YL. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol Breeding. 2010;25:1–12. doi: 10.1007/s11032-009-9323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson LJ, Pearson L, Pearson CJ. Sustainable urban agriculture: stocktake and opportunities. Int J Agr Sustain. 2010;8:7–19. doi: 10.3763/ijas.2009.0468. [DOI] [Google Scholar]
- Pesaresi P, Mizzotti C, Colombo M, Masiero S. Genetic regulation and structural changes during tomato fruit development and ripening. Front Plant Sci. 2014;5:124. doi: 10.3389/fpls.2014.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan P, Fischer G, van Velthuizen H, Reusser DE, Kropp JP. Closing yield gaps: how sustainable can we be? PLoS ONE. 2015;10:e0129487. doi: 10.1371/journal.pone.0129487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017;171:470–480. doi: 10.1016/j.cell.2017.08.030. [DOI] [PubMed] [Google Scholar]
- Santillan Martinez MI, Bracuto V, Koseoglou E, Appiano M, Jacobsen E, Visser RGF, Wolters AA, Bai Y. CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 2020;20:284. doi: 10.1186/s12870-020-02497-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Tabata S, Hirakawa H, Asamizu E, Shirasawa K, Isobe S, Kaneko T, Nakamura Y, et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilbert HM, Rempel A, Pucker B. Comparison of read mapping and variant calling tools for the analysis of plant NGS data. Plant. 2020;9:439. doi: 10.3390/plants9040439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour GB, Ostergaard L, Chapman NH, Knapp S, Martin C. Fruit development and ripening. Annu Rev Plant Biol. 2013;64:219–241. doi: 10.1146/annurev-arplant-050312-120057. [DOI] [PubMed] [Google Scholar]
- Sheehy RE, Kramer M, Hiatt WR. Reduction of polygalacturonase activity in tomato fruit by antisense Rna. Proc Natl Acad Sci USA. 1988;85:8805–8809. doi: 10.1073/pnas.85.23.8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T, Kondo A. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35:441–443. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- Smith CJS, Watson CF, Morris PC, Bird CR, Seymour GB, Gray JE, Arnold C, Tucker GA, Schuch W, Harding S, Grierson D. Inheritance and effect on ripening of antisense polygalacturonase genes in transgenic tomatoes. Plant Mol Biol. 1990;14:369–379. doi: 10.1007/BF00028773. [DOI] [PubMed] [Google Scholar]
- Smith CJS, Watson CF, Ray J, Bird CR, Morris PC, Schuch W, Grierson D. Antisense Rna inhibition of polygalacturonase gene-expression in transgenic tomatoes. Nature. 1988;334:724–726. doi: 10.1038/334724a0. [DOI] [Google Scholar]
- Smith DL, Abbott JA, Gross KC. Down-regulation of tomato beta-galactosidase 4 results in decreased fruit softening. Plant Physiol. 2002;129:1755–1762. doi: 10.1104/pp.011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subburaj S, Chung SJ, Lee C, Ryu SM, Kim DH, Kim JS, Bae S, Lee GJ. Site-directed mutagenesis in Petunia x hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 2016;35:1535–1544. doi: 10.1007/s00299-016-1937-7. [DOI] [PubMed] [Google Scholar]
- Sun KL, van Tuinen A, van Kan JAL, Wolters AMA, Jacobsen E, Visser RGF, Bai YL. Silencing of DND1 in potato and tomato impedes conidial germination, attachment and hyphal growth of Botrytis cinerea. BMC Plant Biol. 2017;17:235. doi: 10.1186/s12870-017-1184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitashev S, Schwartz C, Lenderts B, Young JK, Cigan AM. Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun. 2016;77:13274. doi: 10.1038/ncomms13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitashev S, Young JK, Schwartz C, Gao HR, Falco SC, Cigan AM. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015;169:931–945. doi: 10.1104/pp.15.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama M, Ezura H. How and why does tomato accumulate a large amount of GABA in the fruit? Front Plant Sci. 2015;6:612. doi: 10.3389/fpls.2015.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng XL, Voytas DF, Zhong ZH, Chen YY, Ren QR, Li Q, Kirkland ER, Zhang Y, Qi YP. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants. 2017;3:17018. doi: 10.1038/nplants.2017.18. [DOI] [PubMed] [Google Scholar]
- Tang X, Ren QR, Yang LJ, Bao Y, Zhong ZH, He Y, Liu SS, Qi CY, Liu BL, Wang Y, Sretenovic S, Zhang YX, Zheng XL, Zhang T, Qi YP, Zhang Y. Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol J. 2019;17:1431–1445. doi: 10.1111/pbi.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Sretenovic S, Ren QR, Jia XY, Li MK, Fan TT, Yin DS, Xiang SY, Guo YH, Liu L, Zheng XL, Qi YP, Zhang Y. Plant prime editors enable precise gene editing in rice cells. Mol Plant. 2020;13:667–670. doi: 10.1016/j.molp.2020.03.010. [DOI] [PubMed] [Google Scholar]
- Tashkandi M, Ali Z, Aljedaani F, Shami A, Mahfouz MM. Engineering resistance against tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal Behav. 2018;13:e1525996. doi: 10.1080/15592324.2018.1525996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Handa AK. Reduction in pectin methylesterase activity modifies tissue integrity and cation levels in ripening tomato (Lycopersicon-Esculentum Mill) fruits. Plant Physiol. 1994;106:429–436. doi: 10.1104/pp.106.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Harriman RW, Ramamohan G, Handa AK. An antisense pectin methylesterase gene alters pectin chemistry and soluble solids in tomato fruit. Plant Cell. 1992;4:667–679. doi: 10.2307/3869525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D, Zhu G, Resende MF, Jr, Lin T, Nguyen C, Bies D, Rambla JL, Beltran KS, Taylor M, Zhang B, Ikeda H, Liu Z, Fisher J, Zemach I, Monforte A, Zamir D, Granell A, Kirst M, Huang S, Klee H. A chemical genetic roadmap to improved tomato flavor. Science. 2017;355:391–394. doi: 10.1126/science.aal1556. [DOI] [PubMed] [Google Scholar]
- Tigchelaar E. A new ripening mutant, non-ripening (nor) Rep Tomato Genet Coop. 1973;35:20. [Google Scholar]
- Tiwari M, Trivedi PK, Pandey A. Emerging tools and paradigm shift of gene editing in cereals, fruits, and horticultural crops for enhancing nutritional value and food security. Food Energy Secur. 2020 doi: 10.1002/fes3.258. [DOI] [Google Scholar]
- Ueta R, Abe C, Watanabe T, Sugano SS, Ishihara R, Ezura H, Osakabe Y, Osakabe K. Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci Rep. 2017;7:507. doi: 10.1038/s41598-017-00501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uluisik S, Chapman NH, Smith R, Poole M, Adams G, Gillis RB, Besong TM, Sheldon J, Stiegelmeyer S, Perez L, Samsulrizal N, Wang D, Fisk ID, Yang N, Baxter C, Rickett D, Fray R, Blanco-Ulate B, Powell AL, Harding SE, Craigon J, Rose JK, Fich EA, Sun L, Domozych DS, Fraser PD, Tucker GA, Grierson D, Seymour GB. Genetic improvement of tomato by targeted control of fruit softening. Nat Biotechnol. 2016;34:950–952. doi: 10.1038/nbt.3602. [DOI] [PubMed] [Google Scholar]
- van de Wouw M, Kik C, van Hintum T, van Treuren R, Visser B. Genetic erosion in crops: concept, research results and challenges. Plant Genet Resour-C. 2010;8:1–15. doi: 10.1017/S1479262109990062. [DOI] [Google Scholar]
- Veillet F, Perrot L, Chauvin L, Kermarrec MP, Guyon-Debast A, Chauvin JE, Nogue F, Mazier M. Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int J Mol Sci. 2019;20:402. doi: 10.3390/ijms20020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Somerville S. Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. P Natl Acad Sci USA. 2000;97:1897–1902. doi: 10.1073/pnas.030531997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science. 2002;296:343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- Vu TV, Sivankalyani V, Kim EJ, Doan DTH, Tran MT, Kim J, Sung YW, Park M, Kang YJ, Kim JY. Highly efficient homology-directed repair using CRISPR/Cpf1-geminiviral replicon in tomato. Plant Biotechnol J. 2020;18:2133–2143. doi: 10.1111/pbi.13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AM. Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu Rev Phytopathol. 2015;53:45–66. doi: 10.1146/annurev-phyto-080614-120001. [DOI] [PubMed] [Google Scholar]
- Wang F, Wang C, Liu P, Lei C, Hao W, Gao Y, Liu YG, Zhao K. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE. 2016;11:e0154027. doi: 10.1371/journal.pone.0154027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MG, Mao YF, Lu YM, Wang ZD, Tao XP, Zhu JK. Multiplex gene editing in rice with simplified CRISPR-Cpf1 and CRISPR-Cas9 systems. J Integr Plant Biol. 2018;60:626–631. doi: 10.1111/jipb.12667. [DOI] [PubMed] [Google Scholar]
- Wang DD, Samsulrizal NH, Yang C, Allcock NS, Craigon J, Blanco-Ulate B, Ortega-Salazar I, Marcus SE, Bagheri HM, Perez-Fons L, Fraser PD, Foster T, Fray R, Knox JP, Seymour GB. Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 2019;179:544–557. doi: 10.1104/pp.18.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RF, Tavano ECD, Lammers M, Martinelli AP, Angenent GC, de Maagd RA. Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci Rep. 2019;9:1696. doi: 10.1038/s41598-018-38170-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zhang HY, Zhu HL. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic Res. 2019;6:77. doi: 10.1038/s41438-019-0159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu J, Lao K, Wang S, Mo X, Xu X, Chen X, Mo B. Increasing the efficiency of CRISPR/Cas9-based gene editing by suppressing RNAi in plants. Sci China Life Sci. 2019;62:982–984. doi: 10.1007/s11427-019-9514-9. [DOI] [PubMed] [Google Scholar]
- Wang R, Angenent GC, Seymour G, de Maagd RA. Revisiting the role of master regulators in tomato ripening. Trends Plant Sci. 2020;25:291–301. doi: 10.1016/j.tplants.2019.11.005. [DOI] [PubMed] [Google Scholar]
- Wang SX, Zong Y, Lin QP, Zhang HW, Chai ZZ, Zhang DD, Chen KL, Qiu JL, Gao CX. Precise, predictable multi-nucleotide deletions in rice and wheat using APOBEC-Cas9. Nat Biotechnol. 2020;38:1460–1465. doi: 10.1038/s41587-020-0566-4. [DOI] [PubMed] [Google Scholar]
- Wang WQ, Allan AC, Yin XR. Small RNAs with a big impact on horticultural traits. Crit Rev Plant Sci. 2020;39:30–43. doi: 10.1080/07352689.2020.1741923. [DOI] [Google Scholar]
- Wen B, Strom A, Tasker A, West G, Tucker GA. Effect of silencing the two major tomato fruit pectin methylesterase isoforms on cell wall pectin metabolism. Plant Biol. 2013;15:1025–1032. doi: 10.1111/j.1438-8677.2012.00714.x. [DOI] [PubMed] [Google Scholar]
- White PJ. Recent advances in fruit development and ripening: an overview. J Exp Bot. 2002;53:1995–2000. doi: 10.1093/jxb/erf105. [DOI] [PubMed] [Google Scholar]
- Williamson B, Tudzynsk B, Tudzynski P, van Kan JAL. Botrytis cinerea: the cause of grey mould disease. Mol Plant Pathol. 2007;8:561–580. doi: 10.1111/j.1364-3703.2007.00417.x. [DOI] [PubMed] [Google Scholar]
- Wolabu TW, Cong L, Park JJ, Bao Q, Chen M, Sun J, Xu B, Ge Y, Chai M, Liu Z, Wang ZY. Development of a highly efficient multiplex genome editing system in outcrossing tetraploid Alfalfa (Medicago sativa) Front Plant Sci. 2020;11:1063. doi: 10.3389/fpls.2020.01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JW, Kim J, Il Kwon S, Corvalan C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33:1162–1164. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- Xiao J, Cheng HT, Li XH, Xiao JH, Xu CG, Wang SP. Rice WRKY13 regulates cross talk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements. Plant Physiol. 2013;163:1868–1882. doi: 10.1104/pp.113.226019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GY, Uan MY, Ai CR, Liu LJ, Zhuang E, Karapetyan S, Wang S, Dong XN. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature. 2017;545:491–494. doi: 10.1038/nature22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JM, Hua K, Lang ZB. Genome editing for horticultural crop improvement. Hortic Res. 2019;6:113. doi: 10.1038/s41438-019-0196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Li J, Liu X, Shan T, Qin R, Wei P. Development of plant prime-editing systems for precise genome editing. Plant Commun. 2020;1:100043. doi: 10.1016/j.xplc.2020.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Zhang CW, Yang YX, Zhao S, Kang GT, He XQ, Song JL, Yang JX. Versatile nucleotides substitution in plant using an improved prime editing system. Mol Plant. 2020;13:675–678. doi: 10.1016/j.molp.2020.03.012. [DOI] [PubMed] [Google Scholar]
- Yang L, Huang W, Xiong FJ, Xian ZQ, Su DD, Ren MZ, Li ZG. Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol J. 2017;15:1544–1555. doi: 10.1111/pbi.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TX, Deng L, Zhao W, Zhang RX, Jiang HL, Ye ZB, Li CB, Li CY. Rapid breeding of pink-fruited tomato hybrids using the CRISPR/Cas9 system. J Genet Genomics. 2019;46:505–508. doi: 10.1016/j.jgg.2019.10.002. [DOI] [PubMed] [Google Scholar]
- Yang QH, Wan XS, Wang JY, Zhang YY, Zhang JH, Wang TT, Yang CX, Ye ZB. The loss of function of HEL, which encodes a cellulose synthase interactive protein, causes helical and vine-like growth of tomato. Hortic Res. 2020;7:180. doi: 10.1038/s41438-020-00402-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KQ, Gao CX, Qiu JL. Progress and prospects in plant genome editing. Nat Plants. 2017;3:17107. doi: 10.1038/nplants.2017.107. [DOI] [PubMed] [Google Scholar]
- Yoon YJ, Venkatesh J, Lee JH, Kim J, Lee HE, Kim DS, Kang BC. Genome editing of eIF4E1 in tomato confers resistance to pepper mottle virus. Front Plant Sci. 2020;11:1098. doi: 10.3389/fpls.2020.01098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Li J. Short and long term challenges in crop Breeding. Natl Sci Rev. 2021 doi: 10.1093/nsr/nwab002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Zhao Y. Fluorescence marker-assisted isolation of Cas9-free and CRISPR-edited Arabidopsis plants. Methods Mol Biol. 2019;1917:147–154. doi: 10.1007/978-1-4939-8991-1_11. [DOI] [PubMed] [Google Scholar]
- Yu Q, Han HP, Vila-Aiub MM, Powles SB. AHAS herbicide resistance endowing mutations: effect on AHAS functionality and plant growth. J Exp Bot. 2010;61:3925–3934. doi: 10.1093/jxb/erq205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QH, Wang B, Li N, Tang Y, Yang S, Yang T, Xu J, Guo C, Yan P, Wang Q, Asmutola P. CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci Rep. 2017;7:11874. doi: 10.1038/s41598-017-12262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Hu X, Zhu Y, Mao D. Re-evaluation of the rice ‘Green Revolution’gene: the weak allele SD1-EQ from japonica rice may be beneficial for super indica rice breeding in the post-Green Revolution era. Mol Breeding. 2020;40:1–12. doi: 10.1007/s11032-020-01164-2. [DOI] [Google Scholar]
- Yu H, Lin T, Meng X, Du H, Zhang J, Liu G, Chen M, Jing Y, Kou L, Li X, Gao Q, Liang Y, Liu X, Fan Z, Liang Y, Cheng Z, Chen M, Tian Z, Wang Y, Chu C, Zuo WJJ, Qian Q, Han B, Zuccolo A, Wing R, Gao C, Liang C, Li J. A route to de novo domestication of wild allotetraploid rice. Cell. 2021 doi: 10.1016/j.cell.2021.01.013. [DOI] [PubMed] [Google Scholar]
- Zaidi SSEA, Mukhtar MS, Mansoor S. Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 2018;36:898–906. doi: 10.1016/j.tibtech.2018.04.005. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liang Z, Zong Y, Wang YP, Liu JX, Chen KL, Qiu JL, Gao CX. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun. 2016;7:1–8. doi: 10.1038/ncomms12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Massel K, Godwin ID, Gao CX. Applications and potential of genome editing in crop improvement. Genome Biol. 2018;19:210. doi: 10.1186/s13059-018-1586-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Wu A, Yue Y, Zhao Y. uORFs: important Cis-regulatory elements in plants. Int J Mol Sci. 2020;21:6238. doi: 10.3390/ijms21176238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Appiano M, Pavan S, Bracuto V, Ricciardi L, Visser RG, Wolters AM, Bai Y. Genome-wide study of the tomato SlMLO gene family and its functional characterization in response to the powdery mildew fungus Oidium neolycopersici. Front Plant Sci. 2016;7:380. doi: 10.3389/fpls.2016.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Sretenovic S, Ren Q, Yang L, Bao Y, Qi C, Yuan M, He Y, Liu S, Liu X, Wang J, Huang L, Wang Y, Baby D, Wang D, Zhang T, Qi Y, Zhang Y. Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol Plant. 2019;12:1027–1036. doi: 10.1016/j.molp.2019.03.011. [DOI] [PubMed] [Google Scholar]
- Zhou JP, Deng KJ, Cheng Y, Zhong ZH, Tian L, Tang X, Tang AT, Zheng XL, Zhang T, Qi YP, Zhang Y. CRISPR-Cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front Plant Sci. 2017;8:1598. doi: 10.3389/fpls.2017.01598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yu XQ, Ottosen CO, Rosenqvist E, Zhao LP, Wang YL, Yu WG, Zhao TM, Wu Z. Drought stress had a predominant effect over heat stress on three tomato cultivars subjected to combined stress. BMC Plant Biol. 2017;17:24. doi: 10.1186/s12870-017-0974-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu GT, Wang SC, Huang ZJ, Zhang SB, Liao QG, Zhang CZ, Lin T, Qin M, Peng M, Yang CK, Cao X, Han X, Wang XX, van der Knaap E, Zhang ZH, Cui X, Klee H, Fernie AR, Luo J, Huang SW. Rewiring of the fruit metabolome in tomato breeding. Cell. 2018;172:249–261. doi: 10.1016/j.cell.2017.12.019. [DOI] [PubMed] [Google Scholar]
- Zhu L, Qian Q. Gain-of-function mutations: key tools for modifying or designing novel proteins in plant molecular engineering. J Exp Bot. 2020;71:1203–1205. doi: 10.1093/jxb/erz519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsogon A, Cermak T, Voytas D, Peres LEP. Genome editing as a tool to achieve the crop ideotype and de novo domestication of wild relatives: case study in tomato. Plant Sci. 2017;256:120–130. doi: 10.1016/j.plantsci.2016.12.012. [DOI] [PubMed] [Google Scholar]
- Zsogon A, Cermak T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP. De novo domestication of wild tomato using genome editing. Nat Biotechnol. 2018;36:1211–1216. doi: 10.1038/nbt.4272. [DOI] [PubMed] [Google Scholar]
- Zuriaga E, Blanca JM, Cordero L, Sifres A, Blas-Cerdan WG, Morales R, Nuez F. Genetic and bioclimatic variation in Solanum pimpinellifolium. Genet Resour Crop Ev. 2009;56:39–51. doi: 10.1007/s10722-008-9340-z. [DOI] [Google Scholar]