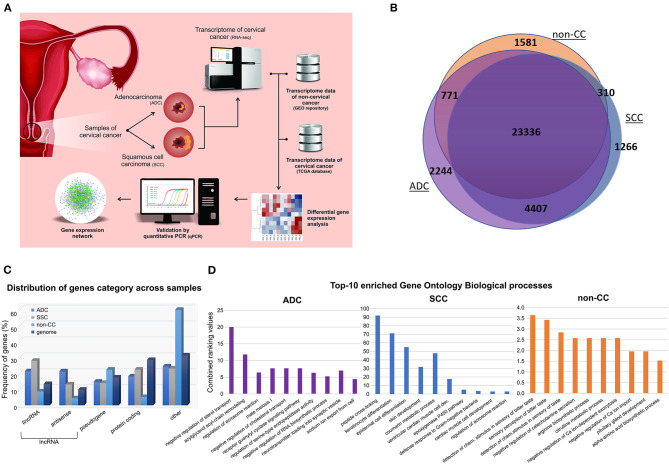
Figure 1.
General design and descriptive results of cervical cancer transcriptome analysis. (A) Pipeline applied in cervical squamous cell carcinoma (SCC) and cervical adenocarcinoma (ADC) samples from our study, followed by inclusion of publicly available HPV-negative non-tumoral cervical tissue (non-CC) samples [10] and cervical SCC and ADC samples from The Cancer Genome Atlas (TCGA). Gene expression of SCC, ADC, and non-CC samples was evaluated. Expression levels of selected long non-coding RNA (lncRNA) genes differentially expressed between cervical ADC and SCC were validated by quantitative real-time PCR. Gene expression networks were predicted using gene expression profiles. (B) Number of genes expressed in SCC and ADC samples from this study and non-CC samples. (C) Frequency of genes expressed in cervical SCC, ADC, and non-CC samples, according to protein-coding, lncRNAs genes, and pseudogenes categories (x-axis). LncRNA genes were subdivided in antisense and long intergenic non-coding RNA (lincRNA), according to Ensembl database annotation (GRCh37/hg19 version). (D) Bar chart of top 10 enriched Gene Ontology biological processes based on gene expression profiles of cervical ADC and SCC samples from our study and non-CC samples, according to combined ranking values (y-axis) calculated by Enrichr (11).