Abstract
Genome-scale mechanistic models of pathways are gaining importance for genomic data interpretation because they provide a natural link between genotype measurements (transcriptomics or genomics data) and the phenotype of the cell (its functional behavior). Moreover, mechanistic models can be used to predict the potential effect of interventions, including drug inhibitions. Here, we present the implementation of a mechanistic model of cell signaling for the interpretation of transcriptomic data as an R/Bioconductor package, a Cytoscape plugin and a web tool with enhanced functionality which includes building interpretable predictors, estimation of the effect of perturbations and assessment of the effect of mutations in complex scenarios.
Keywords: Signaling pathway, Mathematical modelling, Transcriptomic, Causality
1. Introduction
Mechanistic models of signaling pathways provide a natural bridge from variations in genotype (at the scale of gene activity or integrity) to variations in phenotype (at the scale of cells, tissues or organisms) [1]. They are built over graphs that represent the biological knowledge of the complex functional relationships among proteins within the cell, as described in repositories such as KEGG [2], Reactome [3], WikiPathways [4], or other more specialized, such as Disease Maps [5]. Specifically, they provide a conceptual framework for the interpretation of gene expression or genomic variation data and their consequences over downstream processes and phenotypic responses, such as cell proliferation and death, which are particularly relevant for studying disease progression or drug response [6]. Mechanistic models have successfully been used to understand the disease mechanisms behind different cancers [7], [8] (including neuroblastoma [9], [10] and glioblastoma [11]) rare diseases [12], [13], complex diseases such as diabetes [14] or obesity [15], the mechanisms of action of drugs [16] or gender-specific effects of drugs in cancer [17]. In addition to diseases, other scenarios have been studied, such as the molecular mechanisms of death and the post-mortem ischemia of a tissue [18] or the effects of nanoplastics on embryos and human induced pluripotent stem cells [19].
One of the most important aspects of mechanistic models is that they convey the notion of causality and can, therefore, be used to predict the downstream consequences of perturbations of specific conditions [20]. Thus, the possibility of simulating the effect of a drug allowed a systematic in silico drug repurposing experiment in Fanconi Anemia [21] in which some of the drugs predicted were further validated [22]. Also recently, all the targeted drugs currently in clinical trials for testing treatment and prevention of COVID-19 [23] were predicted by means of a mechanistic model [24] of the COVID-19 disease map [25]. Another interesting application of mechanistic modeling show how the simulation of drug inhibitions at single-cell level uncovers the molecular basis of the generation of resistance to bevacizumab in glioblastomas [11]. The effect of mutations can also be simulated with mechanistic models, which demonstrated that mechanistic models are excellent tools to interpret the consequences of complex mutational patterns in diseases such as diabetes [14].
In spite of the usefulness of this innovative approach, the lack of user-friendly bioinformatic tools has hindered the extensive application of mechanistic models to unravel the full potential of interpretation in transcriptomic or genomics experiments. Here we present three implementations of the HiPathia [7] algorithm, a mechanistic model of signaling pathways which provides user-friendly interpretations of the downstream consequences that gene expression levels have over signaling circuits and, ultimately, over cell functionality. In a recent benchmarking, this algorithm has demonstrated to outperform other competing alternatives for modeling signaling pathways [6] (See Supplementary Fig. 1). The HiPathia algorithm is available in three different formats: i) as an R/Bioconductor package for experienced users interested in a programmatic use of the algorithm, as a plugin for the Cytoscape [26] community, which offer a graphic environment for end users, and iii) as a web tool with a dynamic intuitive graphical interface, useful for inexperienced users. The web interface implements extra functionalities beyond the classical differential circuit activity for two-class comparisons, that include the analysis of the impact of simulated interventions (inhibitions, namely knock-outs or knock-downs, over-expressions, etc.) over the activity of the pathways [20] and the evaluation of the potential consequences of mutations over signaling modulation [14]. Moreover, the web interface allows building predictors using signaling circuit activities as features. Interestingly, the features selected by the predictor as relevant for class discrimination provide at the same time valuable insights on the molecular mechanisms that explain the differences between the conditions to discriminate, namely diseases, drug action mechanisms, etc.
2. Material and methods
2.1. The Hipathia mechanistic model
The Hipathia mechanistic model allows modeling the behavior of signaling pathways, described as directed graphs that connect receptor proteins to effector proteins through a chain of activations and inhibitions exerted by intermediate proteins. However, pathways available in the different repositories (KEGG [2], Reactome [3], WikiPathways [4], etc.) describe multifunctional entities with often opposite functionalities (e.g. cell death and cell survival are triggered by the apoptosis pathway, depending on what signaling circuits are transducing the signal). Therefore, to model in detail the specific cell functionalities, any pathway is first decomposed into circuits (Supplementary Fig. 2). Signaling circuits constitute the elementary functional entities that connect one or more receptors to one effector protein, which ultimately triggers specific functions in the cell. The Hipathia algorithm simulates the propagation of the signal considering the level of activity of the proteins that compose the circuit. In this way, different direct (phosphoproteomic) or indirect (protein or gene expression levels) measurements of protein activity can be converted into signaling circuit activities, and consequently, functional profiles of cell activity [7]. In order to be active and therefore transduce the signal to ultimately trigger a function, a circuit requires of the simultaneous presence of the chain of proteins that connect the receptor to the effector, as well the absence of inhibitor proteins that may compromise the transduction of the signal along the circuit. Fig. 1 exemplifies how the signal is transduced across a circuit. In response to specific stimuli (with an arbitrary value of 1), the input node generates signals that are transduced along the pathway following the direction of the interactions until it reaches an output node which triggers some specific cell functionality as response. For each node n within the signal transduction circuit, the signal is propagated along the nodes according to the following recursive rule:
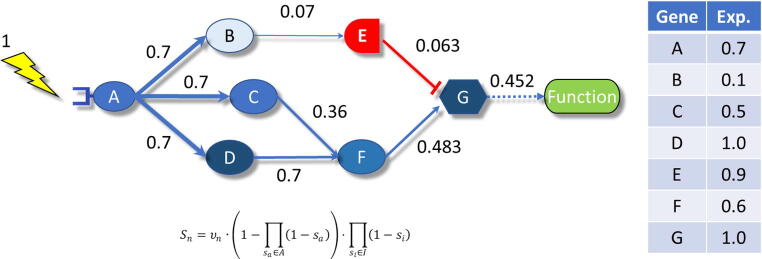
Fig. 1.
Example of a signaling circuit composed by a receptor protein, A, 5 proteins that transduce the signal B, C, D, E and F, one of them being an inhibitor, and an effector protein, G, that ultimately triggers a function. The table represents the normalized gene expression values. The application of the formula below with the values in the table, starting with a hypothetical signal of 1 renders a final intensity signal of 0.452 in the example.
| (1) |
where Sn is the signal intensity for the current node n, vn is its normalized gene expression value, A is the set of activation signals (sa), arriving to the current node from activation edges, Iis the set of inhibitory signals (si) arriving to the node from inhibition edges. See [7] for details. Causality in a holistic context is conveyed by the fact that the expression levels will determine the ultimate functional consequences, following the rules imposed by the signal transduction circuit. Moreover, changes in the activity of the nodes will be reflected (or remain unnoticed) in the last effector node, depending on the topology of each circuit.
2.2. Pathways modelled, user-defined pathways and imported pathways
In the three implementations of the current version of Hipathia more than 8000 circuits have been identified and modeled within a total of more than 150 KEGG [2] pathways of three species (human, mouse and rat).
Also, Simple interaction files (SIF), describing the directional interactions (Activity Flow) among the proteins in the pathway, along with Attributes files (ATT), containing information on the layout of the pathway and formatting details, can be used to build models for new, user-defined pathways using the mgi_from_sif function from the Hipathia Bioconductor package
Thus, pathways from databases that contain Activity Flow structured interactions such as Omnipath [27] or SIGNOR [28], can easily be modelled in Hipathia. Moreover, SIF files can also be extracted from SBML qual files using CaSQ [29], enabling to model pathways from other repositories.
2.3. Circuit functionality annotation
The default Hipathia circuits are annotated using both Uniprot [30] and Gene Ontology (GO) [31] annotations associated with the effector node. This allows obtaining functional profiles in addition to signaling profiles. User-defined or imported pathways can be annotated with the dbannot function from the Hipathia Bioconductor package.
2.4. Predictive model
In addition, we have built a prediction tool that makes discrete class predictors based on circuit signaling activity features, which can be used for class assignation to unknown samples and to augment the interpretability of the data. The model is a pipeline composed of three steps: i) feature selection by means of the Least Absolute Shrinkage and Selection Operator (LASSO) method [32], ii) a Support Vector Machine (SVM) [33] for predicting the class of the samples and iii) a cross-validation operator used to select the best hyperparameters of the methods (the whole procedure is explained with more detail in the web documentation). Thanks to this classification pipeline we can predict the class of unknown samples along with a ranking (using the absolute value of the LASSO coefficients) of which circuits are more important in accordance with the classification problem at hand. The SVM has been implemented using the LIBSVM library [34] by means of the R interface provided by the package e1071 [35]), whereas the LASSO is built with the glmnet R package which implements a fast coordinate descent version of the LASSO [36].
2.5. Implementation
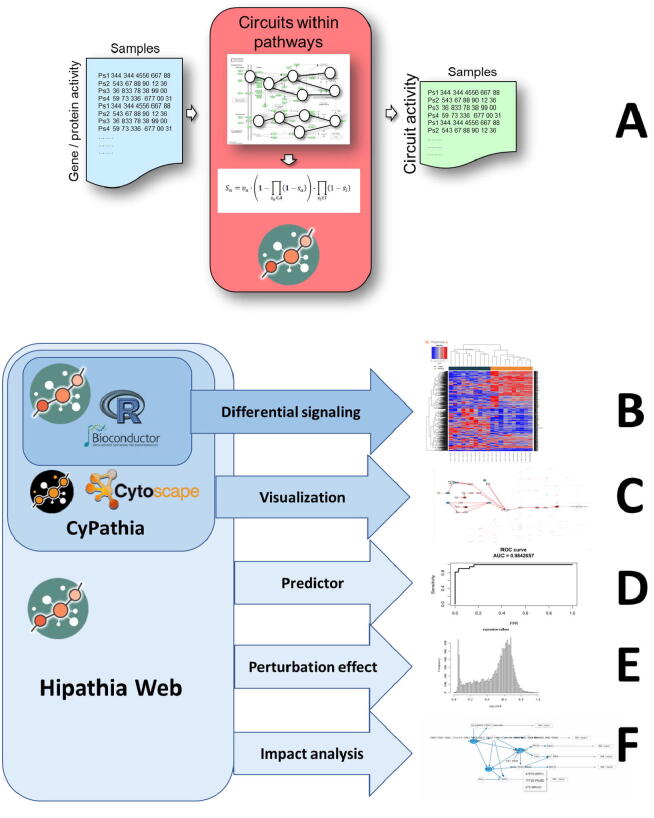
The R/Bioconductor package implements the Hipathia algorithm behind the mechanistic model of cell signaling [7]. The R/Bioconductor Hipathia application implements the model that transforms the gene (or protein) expression matrix into a circuit signaling matrix (Fig. 2A) and only offers the possibility of, given the meta-information that distinguishes two classes within the data, carrying out a differential signaling text (Fig. 2B).
Fig. 2.
HiPathia use and functionalities. A) Transformation of matrices of gene (or protein) expression (activity) into matrices of circuit activities, by decomposing pathways into circuits. B) Estimation of differential signaling, a core functionality included in the R/Bioconductor Hipathia package. C) Interactive visualization of the circuits differentially activated, a functionality implemented in the Cypathia (the Cytoscape implementation, as well as in the web implementations. D) Generation of class predictors based on circuit signaling activity features, and its use for class assignation to unknown samples. E) Estimation of the effect of perturbations (KOs, overexpression, drug inhibitions, etc.) on the circuits and the downstream functionalities. F) Estimation of the impact of mutations in complex systems, considering its downstream effect on cell functionality.
This application is the core for the Cytoscape plugin and the web applications. The Cytoscape plugin, adds the visual representation to the differential signaling test (Fig. 2C).
The web application uses the R/Bioconductor application as backend to implement new functionalities in the front-end. These are: the possibility of obtaining class predictors using circuit activity profiles as features (Fig. 2D), The estimation of the effect of perturbations (e.g. knock-outs, inhibitions, overexpression, drug effects, etc.) on a given condition (Fig. 2E), and estimating the effect of loss-of-function (LoF) mutations in the Impact analysis functionality (Fig. 2F). The client of the web application has been implemented in JavaScript using the HTML5 and SVG standards and uses CellMaps [37] libraries for interactive visual representation of pathways. By default, the web server starts in anonymous mode for occasional use, but it also allows free user registration. Registered users can have permanent storage of their data and analyses done.
2.6. Input data
The Hipathia Bioconductor package, which provides the core functionality to the web interface and the Cytoscape plugin, accepts input data in Comma-separated value (CSV) format with samples as columns and genes/proteins as rows. In order to Hipathia identify gene or protein names, standard IDs must be used. The program can deal with more than 20 standard gene or probe IDs for human, mouse and rat. The data typically comes from gene expression, measured with any sequencing or microarray technology, but also proteomics or phosphoproteomic data can be uploaded. It is assumed that the data has already been normalized for correcting any possible sequencing bias, including batch effect correction. Like in any methodology based on gene expression (e.g. conventional differential gene expression tests) the type of normalization carried out could have an effect of the final results. There are tools and workflows available for the normalization of gene expression data [38]. In the case of proteomic data, the format requirements are the same. Users of different programs like Skyline [39] or MaxQuant [40], or commercial software, like Peaks, ProteomeDiscoverer, etc., have to export the data in text format (CSV) and normalize them.
3. Results and discussion
3.1. Differential signaling activity and visualization of the results
Circuit signaling activity values (Fig. 1A) can be used to conduct tests that compare two conditions or to obtain correlations with a continuous variable (Fig. 1B). Hipathia inputs a matrix of normalized gene (or protein) expression profiles of samples belonging to different classes (defined in the metadata file) and transform it in the corresponding circuit activity matrix (Fig. 1A). This matrix can be used to conduct a test of differential activity between samples belonging to the different groups (Wilcoxon test is used) whose sign indicates up- or down-regulation of the circuits. In addition to discrete classes, a continuous value can be associated to any sample (e.g., the value of a metabolite, a continuous-value phenotype, etc.) Then, a regression can be conducted to detect circuits associated to changes in the value measured for each sample. All the p-values are corrected for multiple testing [41]. The web interface and the Cytoscape plugin provide an intuitive interface to indicate the experimental design (paired or unpaired). select the pathways to be included in the comparison and provide a visual representation of the results as heatmaps and PCA plots (Fig. 1B). An interactive graphical view of the pathways analyzed in which the individual signaling circuits are individualized, highlighting those in which significant changes in signaling was found, along with the downstream functional consequences (Fig. 1C).
The HiPathia documentation presents an interesting example of differential signaling between the estrogen receptor positive (ER+) breast cancer patients versus estrogen negative (ER-) ones. A total of 178 samples of breast cancer tumors were randomly selected from the dataset available in The Cancer Genome Atlas (TCGA) and annotated as ER+ and ER- [42]. Gene expression data of the example was already normalized using trimmed mean of m-values (TMM) [43], and automatically uploaded in the Hipathia web server, where the option Differential signaling activity was chosen.
Gene expression profiles were used to infer signaling activity profiles that were further subjected to a differential activity contrast. As expected, signaling circuits in numerous pathways displayed a different activity between ER+ and ER- samples. Interestingly, most of the circuits of the Estrogen receptor pathway are downregulated in ER-negative tumors (Fig. 3A). Considering that ER-negative tumors are characterized by lacking estrogen receptors, these results are consistent with prior knowledge on the insensitivity to estrogens displayed by this pathway [44].
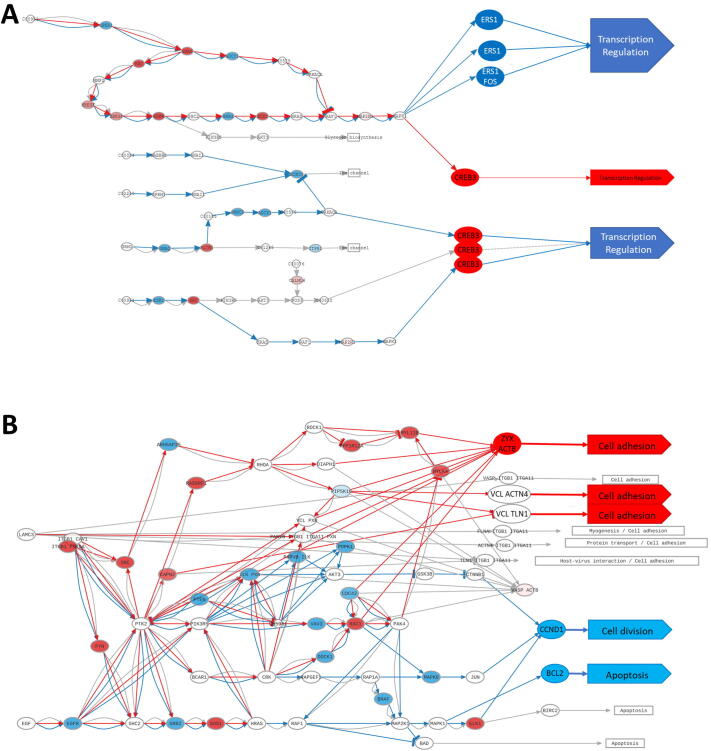
Fig. 3.
Pathways showing signaling circuits differentially activated. Effector nodes and functions affected have been enhanced for clarity. Blue arrows indicate down-activation and red arrows up-activation A) Estrogen receptor (ER) pathway; B) Focal adhesion pathway (hsa04510). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
An interesting aspect of the advantages of holistic approaches, such as the provided by mechanistic models, is illustrated by CREB3 gene that, via several signaling circuits, controls transcription regulation. Despite CREB3 is upregulated in ER-cancers, which in a gene-centric analysis would suggest activation of transcription mediated by estrogens, the holistic analysis of Hipathia ultimately renders a generalized down-activation of the signaling activity in the whole Estrogen receptor pathway due to the down-activation of most of the signaling circuits that activates CREB3 (Fig. 3A).
In the focal adhesion pathway, the activity of proteins involved in actin polymerization, such as actinin, vinculin, and zyxin, is upregulated in ER- tumors. Actin polymerization is involved in cell migration processes required for metastasis, which explains their upregulation given that ER- tumors are known to be more prone to migration and metastasis [45].
This example is available in the Hipathia documentation: http://hipathia.babelomics.org/doc/doku.php?id=worked_example_differential_example.
3.2. Predictors based on signaling features
In addition, signaling circuits can be used to build a predictor of discrete classes (e.g., cancer type, see Fig. 1D). The prediction option allows building a predictor with the well-known SVM algorithm [33] and later using it to predict class membership for unknown samples. Predictors based on gene expression are almost as old as the own transcriptomics. One of the most successful decision support systems in breast cancer, currently known as Mammaprint® in its commercial implementation, was first proposed as early as by 2002 [46]. Since then, many endpoint predictors have been proposed for different cancers [47], [48] and have become mainstream as a decision tool in oncology [49]. Despite the relevance of signaling pathways in the management of cancer is known for long [50] all the available decision support systems based on gene expression use individual genes as features. This hampers the interpretability of the results obtained, given that genes, out of context, cannot be easily related with the functionalities they are ultimately triggering. However, if instead of gene expression levels, the derived signaling circuit activities are used, the interpretability of the result drastically increases as exemplified in a recent predictor of drug sensitivity [16].
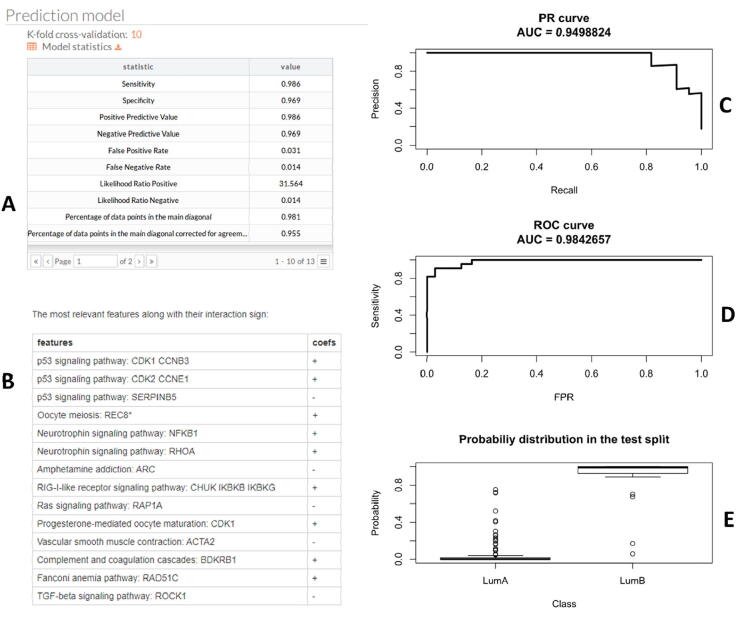
The use of this option can be exemplified by building a predictor to discriminate between luminal A and luminal B breast cancer types, which are relevant for the selection of drug treatment [51]. Again, data is taken from the TCGA breast dataset [42], using first a subset of 505 samples (335 luminal A and 170 luminal B) for training the predictor. The accuracy of the prediction is assessed by cross validation (see the different metrics reported in Fig. 4A). The features that best discriminate between the response values are selected and ranked by means of the LASSO algorithm (see Methods). Interestingly, the first two more relevant features selected in the example are signaling circuits of the P53 signaling pathway (Fig. 4B), with a known central role in breast cancer subtype definition [52]. Finally, the Precision Recall (Fig. 4C) and Receiver Operator (Fig. 4D) curves, along with probability boxplots for the test set (Fig. 4E), for typical train test split scenario are provided. This functionality is available only in the web interface. An example for training and testing the predictor is available in the Hipathia documentation: http://hipathia.babelomics.org/doc/doku.php?id=worked_example_prediction_train_and_test
Fig. 4.
Output of the prediction option. A) Metrics for assessment of the predictor accuracy; B) Most relevant features for the predictor. C) Precision Recall curve; D) Receiver Operator curve; E) probability boxplots.
3.3. Simulation of signaling perturbation experiments
One of the most interesting functionalities of Hipathia is the possibility of simulating the effect of different interventions (e.g. knock-out, over-expression, drug inhibition, etc.) over the activity of signaling circuits in the pathways, as well as their potential downstream functional consequences in the cell [20] (Fig. 1E). Given a specific condition, corresponding to a specific gene expression profile, knock-outs, knock-downs or different types of inhibitions are simulated by reducing or setting to 0 the expression level of the gene or interest. Conversely, over-expressions or agonistic interventions can be simulated by increasing its gene expression level. Additionally, one or several targeted drugs can be selected from a list (taken from DrugBank [53]). The effect of the perturbations is simulated and compared to the original conditions, and the circuits affected are reported.
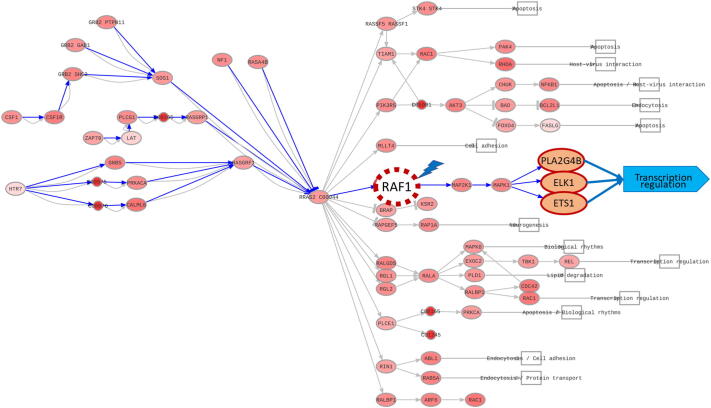
Here, the value of this option is demonstrated by means of an example that focuses on the functional impact that the combined inhibition of EGFR and c-Raf (RAF1) has over pancreatic tumors. In the example, the combined effect of erlotinib (a known and widely used anti-EGFR drug) in combination with the knockdown of the RAF1 gene is simulated. The drug effect is simulated by multiplying the expression value in the pancreatic tumor by the activity factor provided in the settings. To simulate a knockdown, the activity of the target gene is multiplied by the provided knock-down factor. In this example, the effect of Erlotinib reduces EGFR activity to a 10%, while the RAF1 knock-down reduces its activity to a 1% (see RAF1 knock-down effect simulation in Fig. 5). This combination has been described to achieve a complete regression of pancreatic cancer in ductal adenocarcinoma derived xenografts induced by KRAS mutation [54]. This functionality is available only in the web interface. An example for training and testing the predictor is available in the Hipathia documentation: http://hipathia.babelomics.org/doc/doku.php?id=worked_example_perturbation_effect.
Fig. 5.
Perturbation effect. Effect of a knock-down in RAF1 gene over the RAS signaling pathway. Effector nodes and functions affected have been enhanced for clarity.
3.4. Estimation of the impact of mutations on signaling and the downstream cell functionality
Most predictors of the potential pathogenic consequences of mutations are actually only predictors of protein LoF mutations that ignore the actual essentiality of the protein, which is highly dependent on the complex context of interactions with other proteins. Mechanistic models allow estimating the potential pathological role that LoF mutations can have by estimating its effect over the signaling circuits and the downstream functional consequences. This holistic approach, in which the effect of gene mutations is assessed in the context of the rest of variation harbored by the genome, provides an elegant alternative to gene-centric approaches by offering a systems biology perspective for the interpretation of the genomic variability in complex diseases [14]. In this option, the notion of causality implemented in the mechanistic model is again exploited. Knock-outs (KO) are simulated on genes harboring LoF variants in a reference condition (provided by the user or selected from the gene expression profiles of normal tissues available in GTEx [55]). Then the perturbation effect analysis is carried out by comparing the condition with the simulated KOs against the original condition (Fig. 1F).
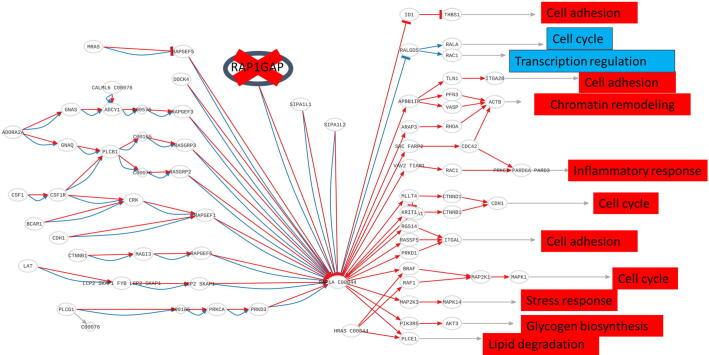
An interesting case study is type 2 diabetes (T2D), a highly prevalent disease (affects over 8% of the population) with a symptomatology that includes several serious comorbilities [56]. Diabetes has a complex and still not completely known genetic background, difficult to assess because the disease phenotype is also highly dependent of environmental factors [57]. To exemplify the use of the Variant interpreter option LoF mutations have systematically been simulated on genes belonging to pathways related to inflammation, one of the most relevant consequences of diabetes [58], over expression profiles of healthy pancreatic donors (taken from GEO repository, ID: GSE38642) [59]. Most of the simulated KOs rendered either non-significant effects on the pathways, or highly significant effect in genes with almost no polymorphisms (and likely lethal and consequently not involved in diabetes) [14]. However, a few of them were suggestive of being involved in diabetes, as the RAP1GAP LoF shown in Fig. 6. Since RAP1GAP is an inhibitor of the Rap1 pathway, the cell functionalities triggered by the circuits that compose the pathway are predicted to be constitutively activated. The functional consequences of the KO of RAP1GAP vary from up-regulation of Inflammatory processes, Cell adhesion and Stress response among others. The results strongly suggest that RAP1GAP plays an important role in the control of diabetes-related processes and therefore, LoF mutations inactivating such protein will most likely result in an over-activation of inflammatory response and thus, an increased risk of developing immune-mediated diabetes [14]. This example can be found in the Hipathia documentation pages: http://hipathia.babelomics.org/doc/doku.php?id=workvafin#results_and_interpretation.
Fig. 6.
Holistic assessment of the impact of mutations. LoF of gene RAP1GAP, an inhibitor of the Rap1 pathway, causes a constitutive activation of different inflammatory processes in the cell. Functions.
4. Discussion
Before highlighting the advantages of mechanistic models for uncovering disease mechanisms or for predicting the consequences of therapeutic interventions is necessary to remark what mechanistic models are not. Mechanistic models are not enrichment analysis methods [60], in which a list of genes selected by any reason (e.g. they were found as differentially expressed in a given experiment) is tested to discover if some functional labels, typically gene ontology terms, are significantly more abundant in them than in the background or in a reference group [61], [62]. Having a significant enrichment of genes labeled as angiogenesis, for example, does not indicate whether these genes are promoting or inhibiting angiogenesis and, moreover, provide no clues on what would be the consequences of interventions over the genes. Mechanistic models are not conventional network analysis methods [63], where a list of genes is interpreted according to their proximity or their connectivity within a known network structure, like the protein–protein-interaction (PPI) network. Although network analysis methods can be an interesting source of hypotheses, the proximity within a biological network does not necessarily imply a causal relationship between the proteins involved. Mechanistic models are built over pathways, which are graphs that represent the detailed knowledge on the way in which cells respond to different stimuli and behave accordingly. Therefore, they implicitly convey the notion of causality, because the activity of the different elements in the model (the genes/proteins) affect the transduction of the signal, accordingly to the pathway topology, and ultimate determine cell behavior. Different methods have been proposed with distinct ways of modeling the transduction of the signal across pathways, such as TAPPA [64], PWEA [65], CLIPPER [66], PRS [67], DEGraph [68], DEAP [69], SubSPIA [70], MinePath [71], or Pathiways [72], [73]. A recent benchmark demonstrated that Hipathia outperformed other alternatives for modeling signaling pathways [6] (See Supplementary Fig. 1).
Also in the context of signaling, other algorithms focus on different types of relationships among proteins, as in the case of CARNIVAL, which focused on regulatory relationships [74]. A different approach is used by tools such as CaSQ, that infers Boolean rules based on the topology and semantics of molecular interaction maps described in SBML [29], or CellCollective that allow collaborative generation of pathway models [75]. However, these tools do allow interpreting transcriptomic data but rather describe the theoretical behavior of the models associated to the underlying pathways.
The data used by the tools and methods above cited are, essentially, gene expression data, taken as proxies of the presence of the activated protein [66], [68], [72], [73], [76], [77]. Obviously, protein expression data can be used instead of gene expression and, in theory, would provide a more accurate approach to the real amount of activated protein. However, other proteomics data types can be used in different ways that are beyond the scope of this type of mechanistic model and obviously beyond the Hipathia functionalities. For example, other types of proteomics applications, such as affinity purification experiments, can be used to infer new protein–protein interaction connections, or posttranscriptional modifications can be used to elucidate cellular signaling events [78], just to cite a few different applications.
Here we provide the core functionality of the implementation of the mechanistic model, along with the most used differential signaling test, implemented in all the tools, and some more functionalities, that are programmatically easy to implement, using the R/Bioconductor Hipathia package, by experienced bioinformaticians, and that have been implemented in the web interface for end users. These include i) the use of signaling activity profiles as “mechanistic” features for building a class predictor, ii) the study of perturbations in a given condition and iii) the assessment of the impact of mutations from a holistic perspective.
The notion of using features that are not the initial measurements (gene expression values) but rather a derived value that accounts for cell functionality of the cell is quite innovative in the field of prediction. Actually, this idea has been recently proposed calling these features dynamic biomarkers or pathway biomarkers [9]. However, a comparative study of the performance of predictors that use signaling profiles versus gene expression profiles across diseases is out of the scope of the manuscript. Probably, in diseases with a strong signaling component, like cancer, they will perform better while in diseases with less representation in the pathways, the use of gene expression will render a better prediction. This tool, simply increases the analytic possibilities of researchers.
In the example presented, the features selected by the LASSO algorithm that best discriminate between the luminal A and B groups contain several interesting signaling circuits. Thus, the two more relevant features are signaling circuits of the P53 signaling pathway (Fig. 4B), known to play a central role in breast cancer subtype definition, as previously commented [52]. Although an exhaustive discussion of the results is out of the scope of this manuscript it is worth mentioning that the rest of circuits selected are relevant in breast cancer, such as Neutrophin signaling pathway, active in carcinomas and target of some therapies [79], or RIG-I-like signaling pathway, also involved in induction of immunogenic response to breast cancer [80] or RAS signaling pathway known to be active in breast cancer [81]. Interestingly, circuits selected by the predictor apparently not related to breast cancer, like the Amphetamine addiction pathway, have already been described as involved in breast cancer therapeutic responses [82].
On the other hand, the option that allows the simulation of signaling perturbation experiments represents an extremely valuable way of predicting the possible effect of drugs (alone or in combination) and other interventions. To demonstrate the value of this functionality of the Hipathia web server, and example of the functional impact that the combined inhibition of EGFR and c-Raf (RAF1) can have on pancreatic tumors was used. This combination has recently been described to produce a complete regression of pancreatic cancer in ductal adenocarcinoma derived xenografts induced by KRAS mutation [54].
The simulation of the combination of the inhibitions suggests a clear impact on cell mechanisms related to several cancer hallmarks, such as proliferation, apoptosis, and cell adhesion and motility. Among the pathways dysregulated by the perturbations, the Ras signaling pathway (KEGG ID: hsa04014), proteoglycans in cancer (hsa05205), ErbB signaling pathway (hsa04012), and PI3K/Akt signaling pathway (hsa04151) are especially remarkable. The dysregulation of these signaling pathways have been described in pancreatic cancer [83] and the in-silico inhibition globally reduces the activity and the functions triggered by these, indeed some targets within them are being used to treat pancreatic tumors (such as EGFR, KRAS, TGFB, NFKB, MEK1). Focusing on the Ras signaling pathway (hsa04014), three circuits appear as inhibited by the knock-down of c-Raf: i) the ELK-1 circuit, which is generally activated in pancreatic cancer [84], ii) the ETS-1 circuit, given that the ETS Proto-Oncogene family (ETS) has been linked to pancreatic cancer progression and, specifically, ETS-1 has been described as potential targets reducing angiogenesis and progression in pancreatic cancer [85], [86], and iii) the PLA2G4B circuit, with the PLA2G4B gene highly expressed in the pancreas, and been this gene described to modulate the sensitivity of pancreatic cancer cells to chemoradiation treatment [87]. Among the most represented effector genes affected by the knock-down simulation, the roles of PLA2G4B, MAPK1, CDKN1A, ELK1, PRKCA, and CAMK2A in processes such as signal transduction, calcium metabolism, and cell cycle regulation must be highlighted. These mechanisms have been highly related to cell proliferation and cancer progression, therefore inactivating the circuits that trigger these effectors would result in a potential reduction of cancer prognosis, consistent with results recently described [54].
5. Conclusions
The mechanistic model implemented in the Hipathia algorithm enables interactive and intuitive evaluation of the impact of gene perturbations on signal transduction from a mechanistic perspective and, consequently, on cell functionality that ultimately determine the phenotype. Here we provide a Bioconductor/R package, along with two user-friendly interfaces, for enabling and facilitating the use of the HiPathia mechanistic model to the scientific community. The web interface implements new functionalities taking advantage of the availability of the core functionality provided by the mechanistic model.
6. Availability
The Bioconductor package is available in the Bioconductor site: https://bioconductor.org/packages/release/bioc/html/hipathia.html.
Documentation on the use of the Hipathia Bioconductor package can be found at: https://bioconductor.org/packages/release/bioc/manuals/hipathia/man/hipathia.pdf
The Cytoscape plugin is available in the Cytoscape site: http://apps.cytoscape.org/apps/cypathia.
The web server is available at: http://hipathia.babelomics.org/
The documentation of the Hipathia web server is available at: http://hipathia.babelomics.org/doc/doku.php
CRediT authorship contribution statement
Kinza Rian: Software. Marta R. Hidalgo: Methodology, Formal analysis. Cankut Çubuk: Formal analysis. Matias M. Falco: Data curation, Validation. Carlos Loucera: Methodology, Formal analysis. Marina Esteban-Medina: Investigation, Data curation. Inmaculada Alamo-Alvarez: . : Investigation, Data curation. María Peña-Chilet: Investigation, Writing - review & editing, Funding acquisition. Joaquín Dopazo: Conceptualization, Supervision, Writing - original draft, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is supported by grants SAF2017-88908-R from the Spanish Ministry of Economy and Competitiveness and PT17/0009/0006 and PI20/01305 from the ISCIII, both co-funded with European Regional Development Funds (ERDF) as well as H2020 Programme of the European Union grants Marie Curie Innovative Training Network “Machine Learning Frontiers in Precision Medicine” (MLFPM) (GA 813533) and “ELIXIR-EXCELERATE fast-track ELIXIR implementation and drive early user exploitation across the life sciences” (GA 676559) to JD. Funding for open access charge: from the Spanish Ministry of Economy and Competitiveness / SAF2017-88908-R.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.05.022.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Yu M.K., Kramer M., Dutkowski J., Srivas R., Licon K., Kreisberg J.F. Translation of genotype to phenotype by a hierarchy of cell subsystems. Cell Syst. 2016;2(2):77–88. doi: 10.1016/j.cels.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucl Acids Res. 2016;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P. The reactome pathway knowledgebase. Nucl Acids Res. 2018;46(D1):D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slenter D.N., Kutmon M., Hanspers K., Riutta A., Windsor J., Nunes N. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018;46(D1):D661–D667. doi: 10.1093/nar/gkx1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostaszewski M., Gebel S., Kuperstein I., Mazein A., Zinovyev A., Dogrusoz U. Community-driven roadmap for integrated disease maps. Briefings Bioinf. 2019;20(2):659–670. doi: 10.1093/bib/bby024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amadoz A., Hidalgo M.R., Çubuk C., Carbonell-Caballero J., Dopazo J. A comparison of mechanistic signaling pathway activity analysis methods. Briefings Bioinf. 2019;20(5):1655–1668. doi: 10.1093/bib/bby040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hidalgo M.R., Cubuk C., Amadoz A., Salavert F., Carbonell-Caballero J., Dopazo J. High throughput estimation of functional cell activities reveals disease mechanisms and predicts relevant clinical outcomes. Oncotarget. 2017;8(3):5160–5178. doi: 10.18632/oncotarget.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cubuk C., Hidalgo M.R., Amadoz A., Pujana M.A., Mateo F., Herranz C. Gene expression integration into pathway modules reveals a pan-cancer metabolic landscape. Cancer Res. 2018;78(21):6059–6072. doi: 10.1158/0008-5472.CAN-17-2705. [DOI] [PubMed] [Google Scholar]
- 9.Fey D., Halasz M., Dreidax D., Kennedy S.P., Hastings J.F., Rauch N. Signaling pathway models as biomarkers: Patient-specific simulations of JNK activity predict the survival of neuroblastoma patients. Sci Signal. 2015;8(408):ra130. doi: 10.1126/scisignal.aab0990. [DOI] [PubMed] [Google Scholar]
- 10.Hidalgo M.R., Amadoz A., Cubuk C., Carbonell-Caballero J., Dopazo J. Models of cell signaling uncover molecular mechanisms of high-risk neuroblastoma and predict disease outcome. Biology Direct. 2018;13(1):16. doi: 10.1186/s13062-018-0219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falco M.M., Peña-Chilet M., Loucera C., Hidalgo M.R., Dopazo J. Mechanistic models of signaling pathways deconvolute the glioblastoma single-cell functional landscape. NAR Cancer. 2020;2(2) doi: 10.1093/narcan/zcaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chacon-Solano E., Leon C., Diaz F., Garcia-Garcia F., Garcia M., Escimez M. Fibroblasts activation and abnormal extracellular matrix remodelling as common hallmarks in three cancer-prone genodermatoses. J Br J Dermatol. 2019;181(3):512–522. doi: 10.1111/bjd.17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.León C., García-García F., Llames S., García-Pérez E., Carretero M., Arriba M.d.C., Dopazo J., del Río M., Escámez M.J., Martínez-Santamaría L. Transcriptomic analysis of a diabetic skin-humanized mouse model dissects molecular pathways underlying the delayed wound healing response. Genes. 2021;12(1):47. doi: 10.3390/genes12010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peña-Chilet M., Esteban-Medina M., Falco M.M., Rian K., Hidalgo M.R., Loucera C. Using mechanistic models for the clinical interpretation of complex genomic variation. Sci Rep. 2019;9(1):1–12. doi: 10.1038/s41598-019-55454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Razzoli M., Frontini A., Gurney A., Mondini E., Cubuk C., Katz L.S. Stress-induced activation of brown adipose tissue prevents obesity in conditions of low adaptive thermogenesis. Molecular metabolism. 2016;5(1):19–33. doi: 10.1016/j.molmet.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amadoz A., Sebastian-Leon P., Vidal E., Salavert F., Dopazo J. Using activation status of signaling pathways as mechanism-based biomarkers to predict drug sensitivity. Sci Rep. 2015;5:18494. doi: 10.1038/srep18494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Çubuk C., Can F.E., Peña-Chilet M., Dopazo J. Mechanistic Models of Signaling Pathways Reveal the Drug Action Mechanisms behind Gender-Specific Gene Expression for Cancer Treatments. Cells. 2020;9(7):1579. doi: 10.3390/cells9071579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira P.G., Muñoz-Aguirre M., Reverter F., Godinho C.P.S., Sousa A., Amadoz A. The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nat Commun. 2018;9(1):490. doi: 10.1038/s41467-017-02772-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bojic S., Falco M.M., Stojkovic P., Ljujic B., Gazdic Jankovic M., Armstrong L. Platform to study intracellular polystyrene nanoplastic pollution and clinical outcomes. Stem Cells. 2020 doi: 10.1002/stem.3244. [DOI] [PubMed] [Google Scholar]
- 20.Salavert F., Hidalgo M.R., Amadoz A., Cubuk C., Medina I., Crespo D. Actionable pathways: interactive discovery of therapeutic targets using signaling pathway models. Nucl Acids Res. 2016;44(W1):W212–W216. doi: 10.1093/nar/gkw369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esteban-Medina M., Peña-Chilet M., Loucera C., Dopazo J. Exploring the druggable space around the Fanconi anemia pathway using machine learning and mechanistic models. BMC Bioinf. 2019;20(1):370. doi: 10.1186/s12859-019-2969-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montanuy H., Martínez-Barriocanal A., Casado J.A., Rovirosa L., Ramírez M.J., Nieto R. Gefitinib and afatinib show potential efficacy for Fanconi anemia-related head and neck cancer. Clin Cancer Res. 2020 doi: 10.1158/1078-0432.CCR-19-1625. [DOI] [PubMed] [Google Scholar]
- 23.Fragkou P.C., Belhadi D., Peiffer-Smadja N., Moschopoulos C.D., Lescure F.-X., Janocha H. Review of trials currently testing treatment and prevention of COVID-19. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loucera C., Esteban-Medina M., Rian K., Falco M.M., Dopazo J., Peña-Chilet M. Drug repurposing for COVID-19 using machine learning and mechanistic models of signal transduction circuits related to SARS-CoV-2 infection. Signal Transduction and Targeted Therapy. 2020;5(1):290. doi: 10.1038/s41392-020-00417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostaszewski M., Mazein A., Gillespie M.E., Kuperstein I., Niarakis A., Hermjakob H. COVID-19 Disease Map, building a computational repository of SARS-CoV-2 virus-host interaction mechanisms. Sci Data. 2020;7(1) doi: 10.1038/s41597-020-0477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Türei D., Korcsmáros T., Saez-Rodriguez J. OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat Methods. 2016;13(12):966–967. doi: 10.1038/nmeth.4077. [DOI] [PubMed] [Google Scholar]
- 28.Licata L., Lo Surdo P., Iannuccelli M., Palma A., Micarelli E., Perfetto L. update. Nucleic Acids Res. 2019;48(D1):D504–D510. doi: 10.1093/nar/gkz949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aghamiri S.S., Singh V., Naldi A., Helikar T., Soliman S., Niarakis A. Automated inference of Boolean models from molecular interaction maps using CaSQ. Bioinformatics. 2020;36(16):4473–4482. doi: 10.1093/bioinformatics/btaa484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.UniProt_Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res 43(Database issue): D204-12. [DOI] [PMC free article] [PubMed]
- 31.Gene_Ontology_Consortium The gene ontology resource: 20 years and still GOing strong. Nucl Acids Res. 2018;47(D1):D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tibshirani R. Regression shrinkage and selection via the lasso. J Roy Stat Soc: Ser B (Methodol) 1996;58(1):267–288. [Google Scholar]
- 33.Vapnik V. John Wiley and Sons; New York: 1999. Statistical Learning Theory. [Google Scholar]
- 34.Chang C.-C., Lin C.-J. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST) 2011;2(3):27. [Google Scholar]
- 35.Meyer D, Dimitriadou E, Hornik K, Weingessel A, Leisch F, Chang C, Lin C. (2014) e1071: Misc functions of the Department of Statistics (e1071), TU Wien. R package version 1(3).
- 36.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1. [PMC free article] [PubMed] [Google Scholar]
- 37.Salavert F., Garcia-Alonso L., Sanchez R., Alonso R., Bleda M., Medina I. Web-based network analysis and visualization using Cell Maps. Bioinformatics. 2016;32(19):3041–3043. doi: 10.1093/bioinformatics/btw332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garrido-Rodriguez M., Lopez-Lopez D., Ortuno F.M., Peña-Chilet M., Muñoz E., Calzado M.A. A versatile workflow to integrate RNA-seq genomic and transcriptomic data into mechanistic models of signaling pathways. PLoS Comput Biol. 2021;17(2) doi: 10.1371/journal.pcbi.1008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 41.Benjamini Y., Yekutieli D. The control of false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 42.TheCancerGenomeAtlasNetwork Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frasor J., Danes J.M., Komm B., Chang K.C., Lyttle C.R., Katzenellenbogen B.S. Profiling of estrogen up-and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144(10):4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 45.Padilla-Rodriguez M., Parker S.S., Adams D.G., Westerling T., Puleo J.I., Watson A.W. The actin cytoskeletal architecture of estrogen receptor positive breast cancer cells suppresses invasion. Nat Commun. 2018;9(1):1–16. doi: 10.1038/s41467-018-05367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van De Vijver M.J., He Y.D., Van't Veer L.J., Dai H., Hart A.A., Voskuil D.W. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 47.Montero-Conde C., Martin-Campos J.M., Lerma E., Gimenez G., Martinez-Guitarte J.L., Combalia N. Molecular profiling related to poor prognosis in thyroid carcinoma. Combining gene expression data and biological information. Oncogene. 2008;27(11):1554–1561. doi: 10.1038/sj.onc.1210792. [DOI] [PubMed] [Google Scholar]
- 48.Buyse M., Loi S., Van't Veer L., Viale G., Delorenzi M., Glas A.M. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98(17):1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 49.Van't Veer L.J., Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452(7187):564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 50.Bild A.H., Yao G., Chang J.T., Wang Q., Potti A., Chasse D. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 51.de Ronde J.J., Hannemann J., Halfwerk H., Mulder L., Straver M.E., Peeters M.-J.T.V. Concordance of clinical and molecular breast cancer subtyping in the context of preoperative chemotherapy response. Breast Cancer Res Treat. 2010;119(1):119–126. doi: 10.1007/s10549-009-0499-6. [DOI] [PubMed] [Google Scholar]
- 52.Bertheau P., Lehmann-Che J., Varna M., Dumay A., Poirot B., Porcher R. p53 in breast cancer subtypes and new insights into response to chemotherapy. The Breast. 2013;22:S27–S29. doi: 10.1016/j.breast.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucl Acids Res. 2017;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blasco MT, Navas C, Martín-Serrano G, Graña-Castro O, Lechuga CG, Martín-Díaz L, Djurec M, Li J, Morales-Cacho L, Esteban-Burgos L. (2019) Complete regression of advanced pancreatic ductal adenocarcinomas upon combined inhibition of EGFR and C-RAF. Cancer Cell 35(4): 573-587. e6. [DOI] [PMC free article] [PubMed]
- 55.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45(6):580. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wild S., Roglic G., Green A., Sicree R., King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27(5):1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 57.Das S.K., Elbein S.C. The Genetic Basis of Type 2 Diabetes. Cellscience. 2006;2(4):100–131. doi: 10.1901/jaba.2006.2-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dandona P., Aljada A., Chaudhuri A., Mohanty P., Garg R. Metabolic syndrome: a comprehensive perspective based on interactions between obesity, diabetes, and inflammation. Circulation. 2005;111(11):1448–1454. doi: 10.1161/01.CIR.0000158483.13093.9D. [DOI] [PubMed] [Google Scholar]
- 59.Taneera J., Lang S., Sharma A., Fadista J., Zhou Y., Ahlqvist E. A Systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab. 2012;16(1):122–134. doi: 10.1016/j.cmet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 60.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucl Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al-Shahrour F., Diaz-Uriarte R., Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20(4):578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 62.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ideker T., Krogan N.J. Differential network biology. Mol Syst Biol. 2012;8:565. doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao S., Wang X. TAPPA: topological analysis of pathway phenotype association. Bioinformatics. 2007;23(22):3100–3102. doi: 10.1093/bioinformatics/btm460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hung J.H., Whitfield T.W., Yang T.H., Hu Z., Weng Z., DeLisi C. Identification of functional modules that correlate with phenotypic difference: the influence of network topology. Genome Biol. 2010;11(2):R23. doi: 10.1186/gb-2010-11-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martini P., Sales G., Massa M.S., Chiogna M., Romualdi C. Along signal paths: an empirical gene set approach exploiting pathway topology. Nucl Acids Res. 2013;41(1) doi: 10.1093/nar/gks866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ibrahim M.A., Jassim S., Cawthorne M.A., Langlands K. A topology-based score for pathway enrichment. J Comput Biol. 2012;19(5):563–573. doi: 10.1089/cmb.2011.0182. [DOI] [PubMed] [Google Scholar]
- 68.Jacob L., Neuvial P., Dudoit S. More power via graph-structured tests for differential expression of gene networks. Ann. Appl. Stat. 2012;6(2):561–600. [Google Scholar]
- 69.Haynes W.A., Higdon R., Stanberry L., Collins D., Kolker E. Differential expression analysis for pathways. PLoS Comput Biol. 2013;9(3) doi: 10.1371/journal.pcbi.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X., Shen L., Shang X., Liu W. Subpathway Analysis based on Signaling-Pathway Impact Analysis of Signaling Pathway. PLoS ONE. 2015;10(7) doi: 10.1371/journal.pone.0132813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koumakis L., Kanterakis A., Kartsaki E., Chatzimina M., Zervakis M., Tsiknakis M. MinePath: mining for phenotype differential sub-paths in molecular pathways. PLoS Comput Biol. 2016;12(11) doi: 10.1371/journal.pcbi.1005187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sebastian-Leon P., Carbonell J., Salavert F., Sanchez R., Medina I., Dopazo J. Inferring the functional effect of gene expression changes in signaling pathways. Nucl Acids Res. 2013;41(Web Server issue):W213–W217. doi: 10.1093/nar/gkt451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sebastian-Leon P., Vidal E., Minguez P., Conesa A., Tarazona S., Amadoz A. Understanding disease mechanisms with models of signaling pathway activities. BMC Syst Biol. 2014;8(1):121. doi: 10.1186/s12918-014-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu A., Trairatphisan P., Gjerga E., Didangelos A., Barratt J., Saez-Rodriguez J. From expression footprints to causal pathways: contextualizing large signaling networks with CARNIVAL. npj Syst Biol Appl. 2019;5(1):1–10. doi: 10.1038/s41540-019-0118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Helikar T., Kowal B., McClenathan S., Bruckner M., Rowley T., Madrahimov A. The cell collective: toward an open and collaborative approach to systems biology. BMC Syst Biol. 2012;6(1):1–14. doi: 10.1186/1752-0509-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Efroni S., Schaefer C.F., Buetow K.H. Identification of key processes underlying cancer phenotypes using biologic pathway analysis. PLoS ONE. 2007;2(5) doi: 10.1371/journal.pone.0000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mitsos A., Melas I.N., Siminelakis P., Chairakaki A.D., Saez-Rodriguez J., Alexopoulos L.G. Identifying drug effects via pathway alterations using an integer linear programming optimization formulation on phosphoproteomic data. PLoS Comput Biol. 2009;5(12) doi: 10.1371/journal.pcbi.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rudolph J.D., Cox J.r. A network module for the Perseus software for computational proteomics facilitates proteome interaction graph analysis. J Proteome Res. 2019;18(5):2052–2064. doi: 10.1021/acs.jproteome.8b00927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hondermarck H. Neurotrophins and their receptors in breast cancer. Cytokine Growth Factor Rev. 2012;23(6):357–365. doi: 10.1016/j.cytogfr.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 80.Elion D.L., Jacobson M.E., Hicks D.J., Rahman B., Sanchez V., Gonzales-Ericsson P.I. Therapeutically active RIG-I agonist induces immunogenic tumor cell killing in breast cancers. Cancer Res. 2018;78(21):6183–6195. doi: 10.1158/0008-5472.CAN-18-0730. [DOI] [PubMed] [Google Scholar]
- 81.von Lintig F.C., Dreilinger A.D., Varki N.M., Wallace A.M., Casteel D.E., Boss G.R. Ras activation in human breast cancer. Breast Cancer Res Treat. 2000;62(1):51–62. doi: 10.1023/a:1006491619920. [DOI] [PubMed] [Google Scholar]
- 82.Wu H., Chen L., Zhu F., Han X., Sun L., Chen K. The cytotoxicity effect of resveratrol: cell cycle arrest and induced apoptosis of breast cancer 4T1 Cells. Toxins. 2019;11(12):731. doi: 10.3390/toxins11120731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iozzo R.V., Sanderson R.D. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011;15(5):1013–1031. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arlt A., Schäfer H., Kalthoff H. The ‘N-factors’ in pancreatic cancer: functional relevance of NF-κB, NFAT and Nrf2 in pancreatic cancer. Oncogenesis. 2012;1(11):e35. doi: 10.1038/oncsis.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lefter L., Dima S., Sunamura M., Furukawa T., Sato Y., Abe M. Transcriptional silencing of ETS-1 efficiently suppresses angiogenesis of pancreatic cancer. Cancer Gene Ther. 2009;16(2):137–148. doi: 10.1038/cgt.2008.65. [DOI] [PubMed] [Google Scholar]
- 86.Kobberup S., Nyeng P., Juhl K., Hutton J., Jensen J. ETS-family genes in pancreatic development. Developmental Dynamics. 2007;236(11):3100–3110. doi: 10.1002/dvdy.21292. [DOI] [PubMed] [Google Scholar]
- 87.Wei D., Parsels L.A., Karnak D., Davis M.A., Parsels J.D., Marsh A.C. Inhibition of protein phosphatase 2A radiosensitizes pancreatic cancers by modulating CDC25C/CDK1 and homologous recombination repair. Clin Cancer Res. 2013;19(16):4422–4432. doi: 10.1158/1078-0432.CCR-13-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.