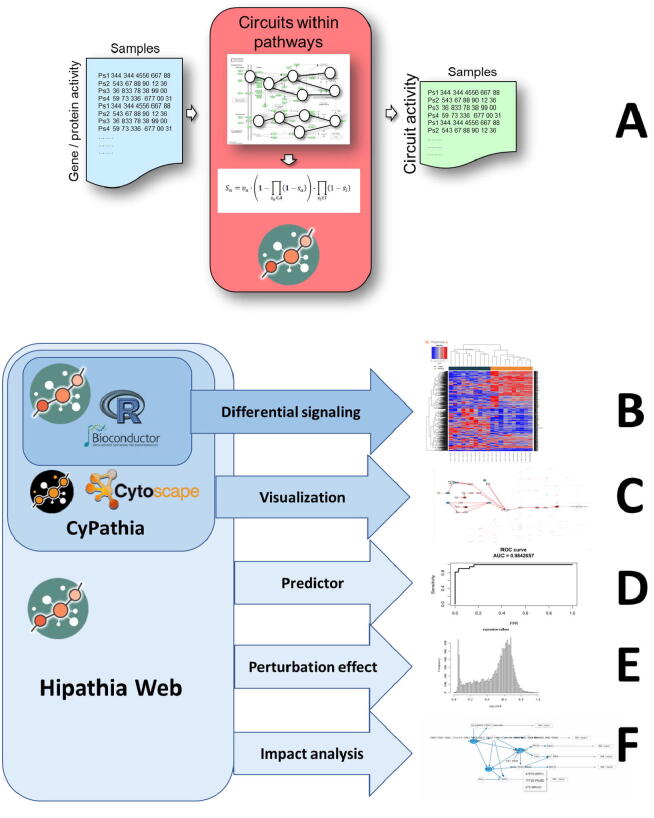
Fig. 2.
HiPathia use and functionalities. A) Transformation of matrices of gene (or protein) expression (activity) into matrices of circuit activities, by decomposing pathways into circuits. B) Estimation of differential signaling, a core functionality included in the R/Bioconductor Hipathia package. C) Interactive visualization of the circuits differentially activated, a functionality implemented in the Cypathia (the Cytoscape implementation, as well as in the web implementations. D) Generation of class predictors based on circuit signaling activity features, and its use for class assignation to unknown samples. E) Estimation of the effect of perturbations (KOs, overexpression, drug inhibitions, etc.) on the circuits and the downstream functionalities. F) Estimation of the impact of mutations in complex systems, considering its downstream effect on cell functionality.