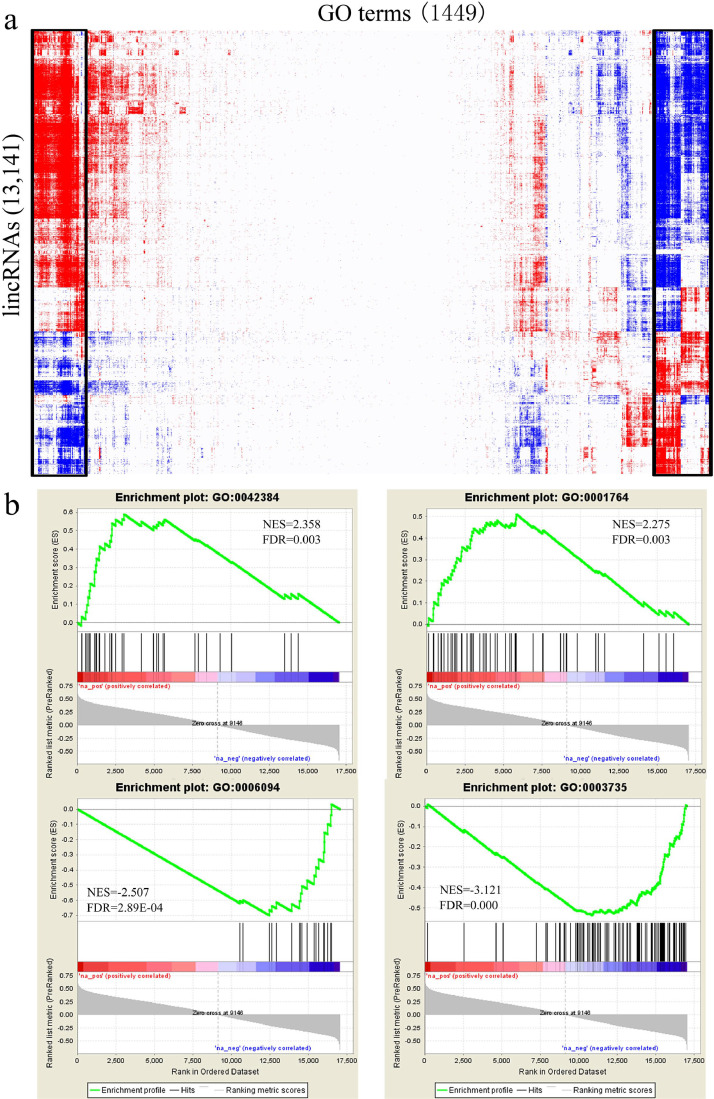
Figure 5.
Functional annotation of lincRNAs: (a) Expression-based association matrix of lincRNAs (rows) and GO terms (columns) derived from GSEA. lincRNAs and GO terms are positively (red; FDR <0.05; NES >0), negatively (blue; FDR <0.05; NES <0), or not associated (white; FDR >0.05). Black boxes highlight significant biclusters in the matrix. (b) GSEA enrichment plots of linc5006 associated with GO:0042384 (cilium assembly), GO:0001764 (neuron migration), GO:0006094 (gluconeogenesis), and GO:0003735 (structural constituent of ribosome). The green curve corresponds to the calculation of enrichment scores (ES). The horizontal bar (red to blue gradient) represents the known ranked genes ordered by Pearson correlation coefficients with linc5006. Vertical black lines represent the projection of individual genes within the tested GO term onto the ranked gene list. Abbreviations: FDR, false discovery rate; lincRNAs, Long noncoding RNAs; NES, normalized enrichment score. (Color version of figure is available online.)