Figure 2.
p97 inhibition results in excessive DNA end resection mediated by MRE11
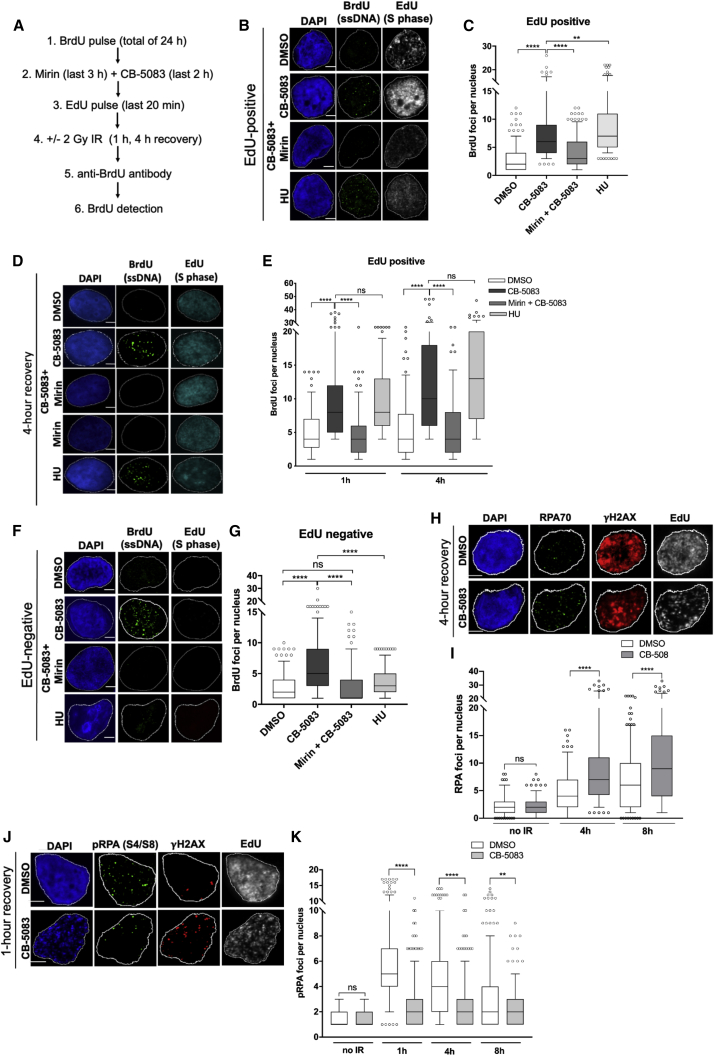
(A) Schematic of the BrdU assay.
(B and C) Representative images (B) and quantification of EdU-positive BrdU foci (C) in CB-5083- and mirin-treated T24 cells. Scale bars, 5 μm; n = 3.
(D and E) Representative images (D) and quantification (E) of BrdU foci in EdU-positive T24 cells after 2 Gy IR and 1 or 4 h of recovery. Scale bars, 5 μm; n = 3.
(F and G) Representative images (F) and quantification (G) of EdU-negative BrdU foci in CB-5083- and mirin-treated T24 cells. Scale bars, 5 μm; n = 3
(H and I) Representative images (H) and quantification (I) of EdU-positive RPA70 foci at 4 h of recovery after 2 Gy IR in CB-5083 treated T24 cells. Scale bars, 5 μm; n = 3.
(J and K) Representative images (J) and quantification (K) of phosphorylated RPA foci (S4/S8) formation at 1 h of recovery from 2 Gy IR. Scale bars, 5 μm; n = 3.
For all graphs, data are presented as boxplot, and values beyond the 5th and 95th percentiles are shown as individual data points. For statistical analysis in (C), (E), and (G), Kruskal-Wallis with Dunn’s multiple comparisons was used. For (I) and (K), the Mann-Whitney test was used. NS, not significant; ∗∗p < 0.01; ∗∗∗∗p < 0.0001. See also Figures S3 and S4.