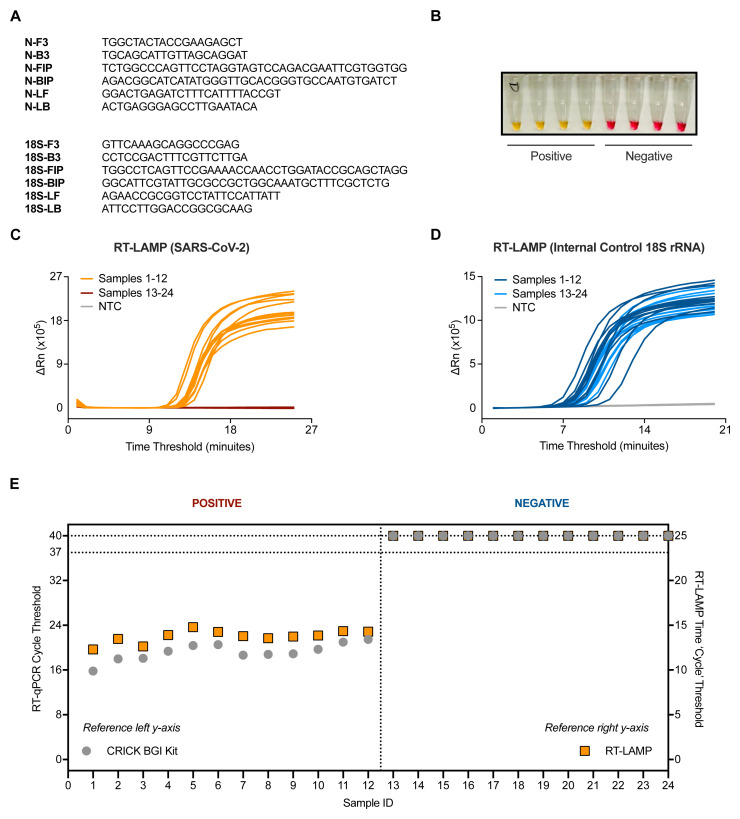
Figure 1. Validation of SARS-CoV-2 detection by RT-LAMP targeting the N gene.
( A) The N gene of SARS-CoV-2 was targeted using primers designed by Zhang et al. 10, whilst the internal control implemented utilised primers for 18S rRNA initially published by Lamb et al. 9. ( B) Upon DNA amplification, a colorimetric pH dye in the reaction mix will change from pink to yellow if the target is present. Four SARS-CoV-2 positive and four SARS-CoV-2 negative patient samples are shown (side view of PCR tubes). ( C–D) Amplification curves from 24 patient samples assessed by SARS-CoV-2 (N gene) RT-LAMP ( C) or 18S ( D) in a Real-Time PCR machine. ( E) Dot plot of the SARS-CoV-2 Ct values from ( C–D) compared to Ct values derived from the Crick’s RT-qPCR diagnostic pipeline using the CE marked BGI kit. Ct values of ‘undetermined’ are plotted as “Ct = 40” (left y-axis) or “Ct = 25” (right y-axis) for illustrative purposes. Data are normalised by assay run/cycles determined by the two methods for comparative purposes. The clinical call from the reference laboratory is indicated above the graph.