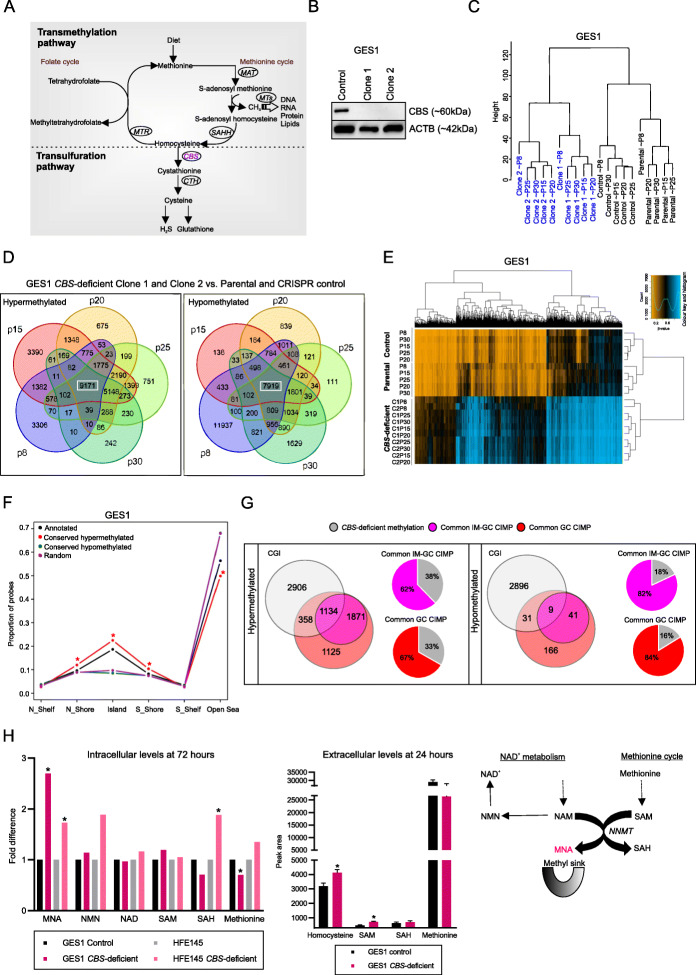
Fig. 4.
Knocking out CBS alters the DNA methylation landscape in normal gastric epithelial cells. a Biochemical position of CBS in the transsulfuration and transmethylation cycles. b CBS protein levels in GES1 CRISPR control and CBS-deficient CRISPR clones. c Cluster dendogram of methylation data indicating branching of GES1 CBS-deficient clones from GES1 parental and CRISPR control cells. d Venn diagram of differential methylation analysis of GES1 CBS-deficient clone 1 and clone 2 compared to parental and CRISPR control cells. e Unsupervised clustering of conserved hypermethylated 9171 CpG sites in GES1 CBS-deficient clones [clone 1, C1; clone 2, C2] across passages. f Breakdown of the GES1 CBS-deficient conserved signature in CpG contexts island, shore, shelf, and open sea; *P < 0.001 according to binomial test; “Annotated” refers to the distribution of CpG probes on the Infinium methylation EPIC array, “Random” refers to the sum of all differentially methylated CpG probes in random pairing of samples. g Comparison of CBS-deficient conserved signatures in GES1 or HFE145 with the “GIM-GC common CIMP” and “GC common CIMP” signatures at CpG islands indicate a larger overlap for hypermethylated (33–38%) events compared to hypomethylation events (16–18%); P < 1 × 10−4, bootstrapping using 10,000 samples. h Graph showing fold difference of intracellular levels of metabolites in CBS-deficient cells (n = 4–5) compared to controls (n = 5) at 72 h, 1-methyl nicotinamide [MNA] (and related metabolites, nicotinamide mononucleotide [NMN], nicotinamide adenine dinucleotide [NAD], S-adenosyl methionine [SAM], S-adenosyl homocysteine [SAH], and methionine, *P < 0.05 (left panel). Graph showing peak area of extracellular levels of homocysteine, SAM, SAH, and methionine in CBS-deficient cells (n = 5) compared to controls (n = 7) at 24 h, *P < 0.05 (middle panel). Biochemical reaction showing conversion of nicotinamide [NAM] to MNA using a methyl group from SAM, catalyzed by nicotinamide N-methyltransferase [NNMT] (right panel). CBS, cystathionine beta-synthase; CTH, cystathionine gamma-lyase; MTR, 5-methyltetrahydrofolate-homocysteine methyltransferase; MAT, Methionine adenosyltransferase; MTs, Methyl transferases