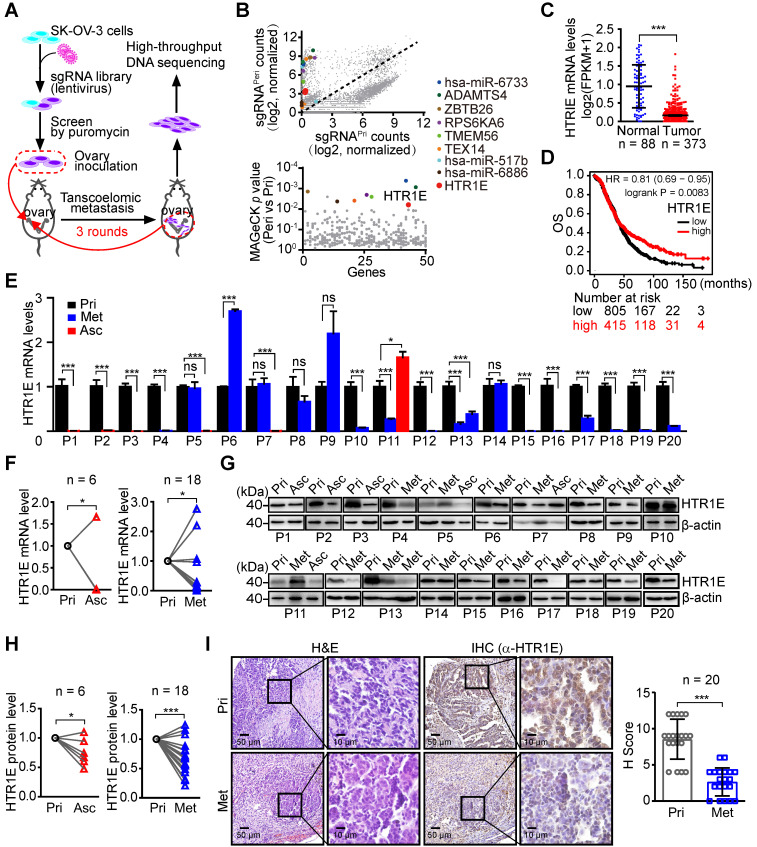
Figure 1.
Genome-wide CRISPR screen identifies HTR1E as a key gene regulating OC peritoneal dissemination. (A) The schematic diagram of genome-wide in vivo screening of key genes regulating peritoneal dissemination by CRISPR/Cas9 library in an OC orthotopic murine model. (B) Scatterplot showing enrichment of specific sgRNAs in peritoneal disseminated (sgRNAPeri) or primary (sgRNAPri) OC xenografts (top panel) and the identification of top candidate genes using MAGeCK P value analysis (bottom panel). (C) The mRNA level of HTR1E in normal human ovarian tissues (n = 88) and OC specimens (n = 373) from Genotype-Tissue Expression (GTEx) database and TCGA database (***P < 0.001, by paired, two-tailed student's t-test). (D) Kaplan-Meier survival curve to show the overall survival (OS) of OC patients with different HTR1E expression (n = 1220). (E-F) qRT-PCR analysis of HTR1E in 20 human OC specimens (P) including 20 primary tumor samples (Pri) with 6 paired ascites samples (Asc) and 17 paired peritoneal OC metastatic nodules (Met) (E) and the statistical analysis on the change of HTR1E expression during OC metastasis (F) (means ± SEM from three independent experiments, *P < 0.05, ns not significant, by paired, two-tailed student's t-test). (G-H) Western blot analysis of HTR1E in human OC primary tumors and paired metastases and ascites from 20 OC patients (G) and the statistical analysis on the change of HTR1E protein during OC metastasis (H) (*P < 0.05, ***P < 0.001, by paired, two-tailed student's t-test). (I) H&E and IHC staining of HTR1E in the primary OC tumors and paired metastases dissected from human OC patients and the quantification by H score method (n = 20, ***P < 0.001, by paired, two-tailed student's t-test).