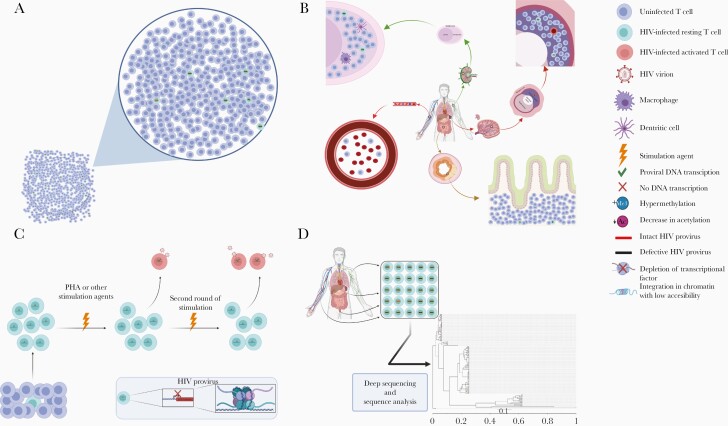
Figure 1.
Challenge of identifying and reactivating human immunodeficiency virus (HIV)–infected cells. A, CD4+ T cells harboring HIV proviral DNA are rare among the total CD4+ T-cell population; most of the proviral DNA is defective, and only <10% is intact. B, HIV-infected cells are mostly located in difficult-to-study anatomic sites, including lymph nodes, intestinal lymphoid tissue, and spleen. C, Multiple potential mechanisms contribute to HIV latency, including epigenetic modifications, depletion of transcriptional factors, and integration in dense regions of the chromosome. Even the most potent CD4+ T-cell–activating agents (eg, phytohemagglutinin [PHA]) are only able to activate a small proportion of CD4+ T cells harboring intact HIV proviruses, and the underlying mechanisms remain elusive. D, HIV achieves sequence diversity very early during infection; sequences obtained from different cell and anatomic compartments demonstrate substantial diversity as shown in this example phylogenetic analysis of intact HIV proviral sequences from 1 participant. Figures were generated with BioRender.com.