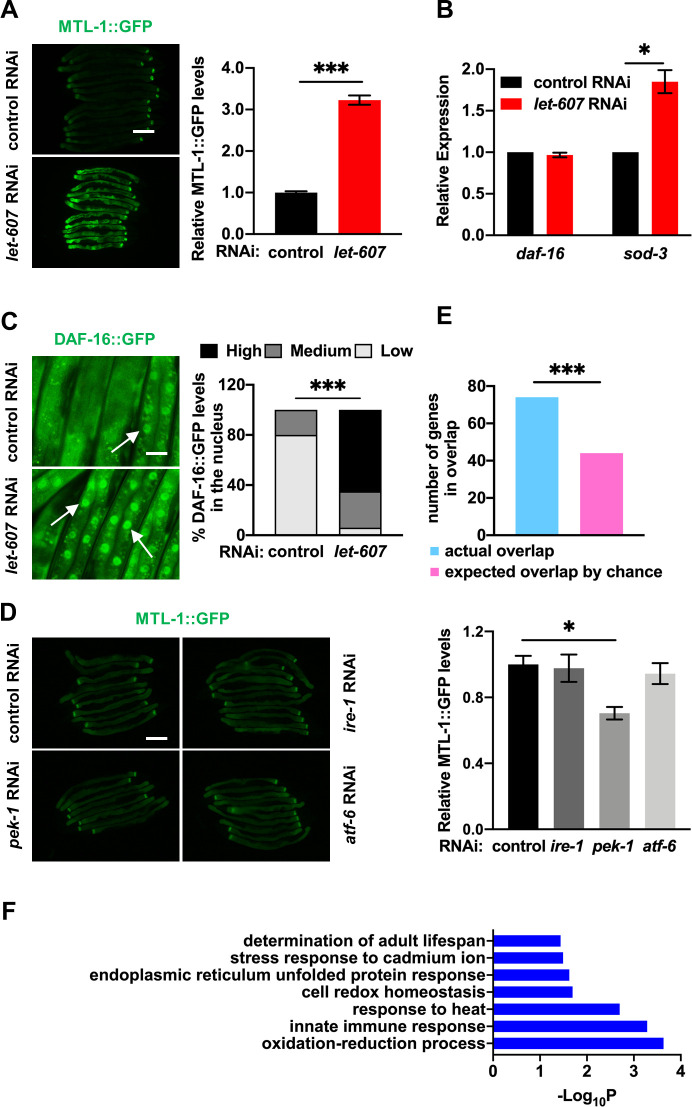
Fig 2. Suppression of LET-607 adaptively activates DAF-16.
(A) Effects of let-607 RNAi on MTL-1::GFP expression. Left panel, representative images. Right panel: quantification data. Scale bar = 100 μm. (B) Effects of let-607 RNAi on the mRNA expression of daf-16 and sod-3 as measured by qPCR. n = 3 per group. (C) Effects of let-607 RNAi on DAF-16::GFP nuclear occupancy. Left panel, representative images. White arrow indicates the nuclear GFP signal. Right panel: semi-quantification data. Scale bar = 25 μm. Number of animals (n): control RNAi (85) and let-607 RNAi (96). (D) RNAi of the ER UPR genes had no effects on MTL-1::GFP expression. Left panel, representative images. Right panel: quantification data. Scale bar = 100 μm. (E) Number of genes that were induced by let-607 RNAi and also DAF-16 class I target genes. *** p < 0.001 versus actual overlap. (F) GO term analysis of let-607 RNAi-induced DAF-16 class I genes. Data were presented as mean ± SEM. * p < 0.05, *** p < 0.001.