Fig. 2. 47D11 binds specifically to the RBD in down conformation and prevents their full compaction.
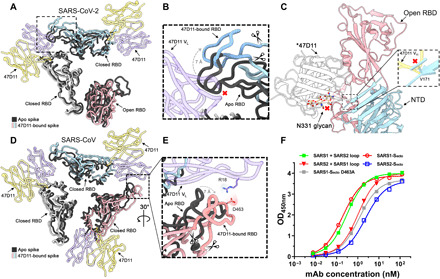
(A) Top view of the 47D11-bound SARS2 spike, shown as a ribbon diagram. The 47D11-bound spike protomers are colored pink, blue, and gray, and the 47D11 HC and LC are shown semitransparently and colored yellow and purple, respectively. Glycans and the NTD are omitted for clarity, and only the Fab variable region is shown. The superposed structure of the partially open apo SARS2 spike (PDB ID: 6ZGG) is colored black. (B) Zoomed-in view of the boxed region in (A). The region encompassing residues 470 to 490, used for the loop swap experiments, is indicated with scissors. (C) Zoomed-in view of the SARS2 up RBD and adjacent NTD, shown in cartoon representation. The overlaid 47D11 Fab is shown as a silhouette, and the N331 glycan is shown in ball-and-stick representation and colored tan. The inset shows a zoomed-in view of the clash between the NTD residue V171 and the 47D11 HC. (D) Top view of the 47D11-bound SARS spike colored as shown in (A). The superposed structure of the closed apo SARS spike (PDB ID: 5XLR) is colored black. (E) Zoomed-in view of the boxed region in (D), showing a putative salt bridge between the 47D11 variable LC and the RBD loop. The region encompassing residues 457 to 477, used for the loop swap experiments, is indicated with scissors. (F) ELISA binding curves of 47D11 binding to wild-type, loop-swapped, and D463A spike ectodomains. VH, variable region of immunoglobulin heavy chain; VL, variable region of immunoglobulin light chain; OD450nm, optical density at 450 nm.
