Fig. 5. Cytidine at position 32 in tRNA-Tyr undergoes C-to-Ψ RNA editing.
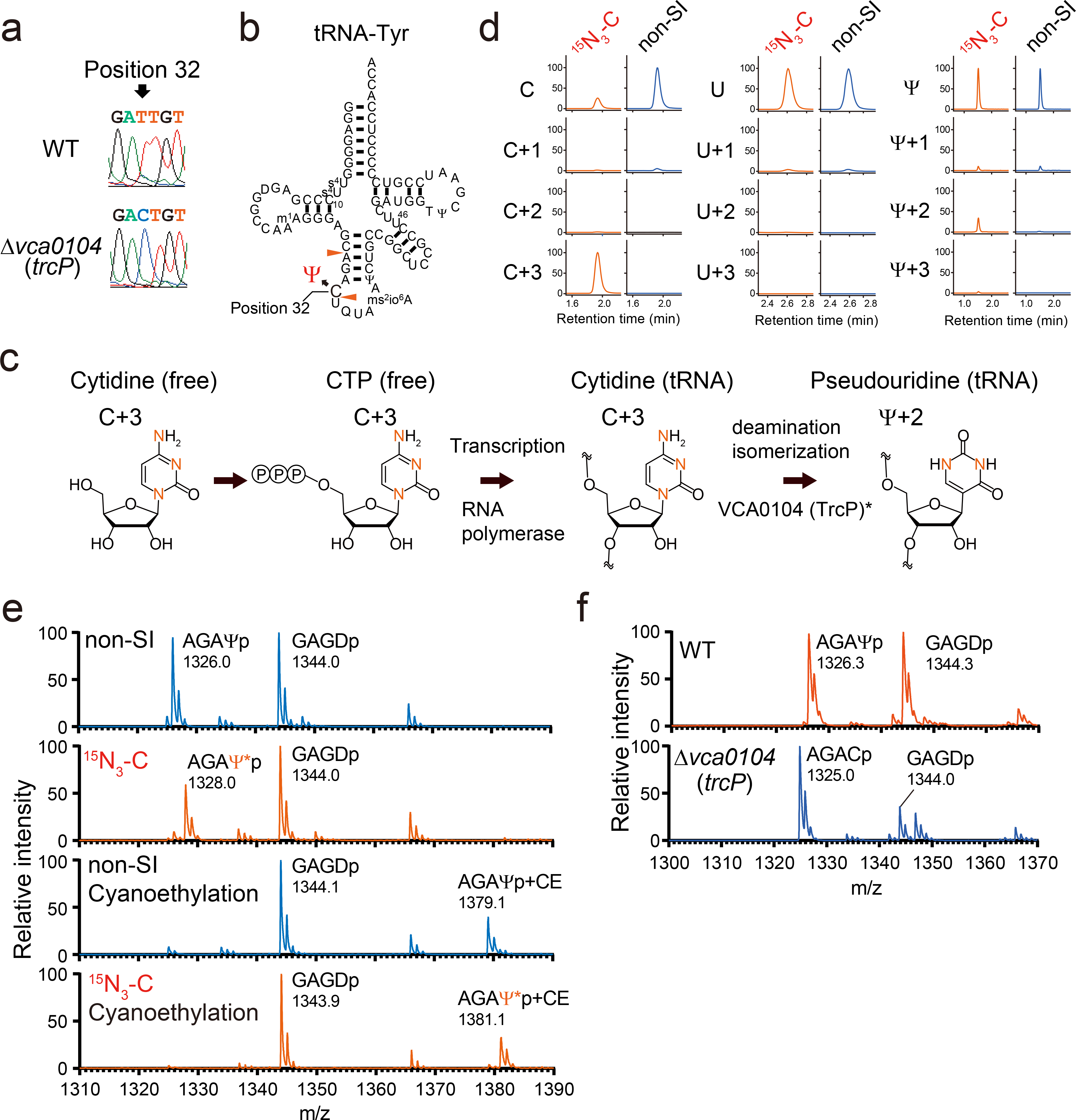
a, Sanger sequencing of cDNA of tRNA-Tyr from WT (top) and Δvca0104 (bottom) strains. Position 32 is indicated by the arrow.
b, Secondary structure of tRNA-Tyr with modifications based on fragment analyses (Extended Data Fig. 2 and Extended Data Fig. 7 and Supplementary Data 3). RNase A cleavage sites that form the fragment containing position 32 are indicated by red arrowheads.
c, Proposed scheme for the conversion of the isotope labeled cytidine to pseudouridine. Red letters represent 15N. *VCA0104 (TrcP) is required for the C-to-Ψ conversion, but its sufficiency for the process has not been established.
d, Nucleoside analysis of purified tRNA-Tyr isolated from Δvc1231, a strain deficient in the conversion of cytidine to uridine and cultured with 15N3-cytidine (15N3-C) or cytidine (non-SI). The detected nucleosides are indicated on the left. Representative data from two independent experiments with similar results is shown.
e, Fragment analyses of an oligo protected portion (position 10 to 46) of tRNA-Tyr isolated from the Δvc1231 strain grown with either cytidine (non-SI) or stable isotope labeled cytidine (15N3-C). In the lower two panels, oligo protected portions were incubated with acrylonitrile, which specifically cyanoethylates (CE) pseudouridine, increasing its mass by 53 Da. m/z values of detected peaks with assigned fragment sequences are shown. Ψ* indicates stable isotope labeled Ψ, which is 2Da heavier. The MALDI analyses were conducted in negative polarity mode. Representative data from two independent experiments with similar results is shown.
f, Fragment analysis of an oligo protected portion (position 10 to 46) of tRNA-Tyr from WT and Δvca0104 (trcP) strains. m/z values of detected peaks with assigned fragment sequences are shown. The MALDI analyses were conducted in negative polarity mode. Representative data from two independent experiments with similar results is shown.
