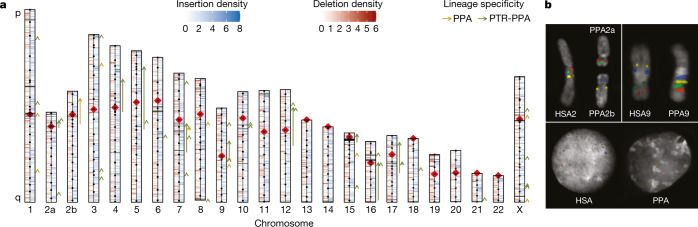
Fig. 1. Sequence and assembly of the bonobo genome.
a, Schematic of the Mhudiblu_PPA_v0 assembly depicting the centromere location (red rhombus), FISH probes used to create assembly backbone (black dots), fixed bonobo-specific insertions (blue) and deletions (red) (Supplementary Data), remaining gaps (black horizontal lines) and large-scale inversions (arrows). We distinguish bonobo-specific inversions (dark orange, PPA) from Pan-specific inversions (dark green, PTR-PPA). b, FISH validation of the bonobo chromosome 2a and 2b fusion and the 2b pericentric inversion (probes: RP11-519H15 in red, RP11-67L14 in green, RP11-1146A22 in blue, RP11-350P7 in yellow) (top left); the chromosome 9 pericentric inversion (probes: RP11-1006E22 in red, RP11-419G16 in green, RP11-876N18 in blue, RP11-791A8 in yellow) (top right); and the inversion Strand-seq_chr7_inv4a (probes: RP11-118D11 in green, WI2-3210F8 in red, RP11-351B3 in blue) (bottom).