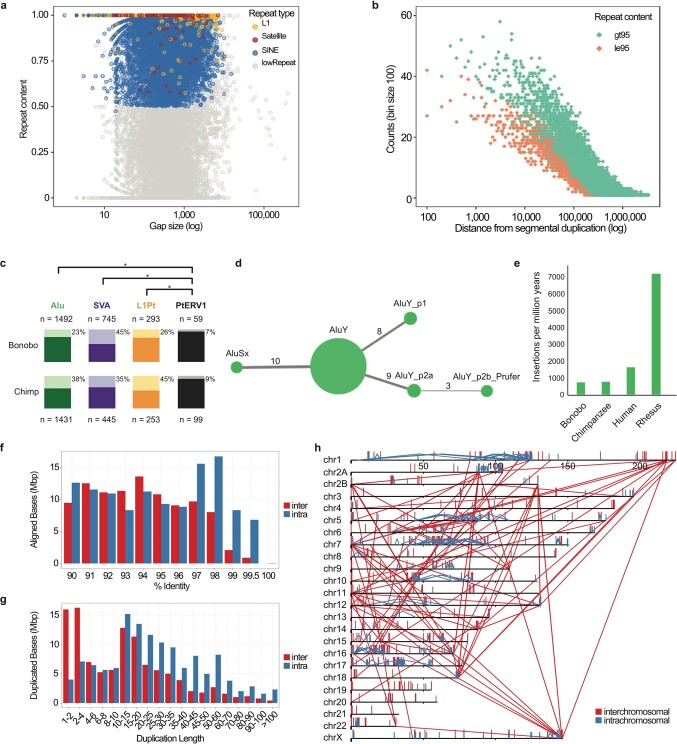
Extended Data Fig. 3. Sequence and assembly of the bonobo genome and bonobo genome repeat structure.
a, The size (x axis is shown on a log scale) and repeat content of gaps filled in the new bonobo assembly compared with the panpan1.1 assembly1. Gaps composed of more than 50% repeat content for any particular class of repeat are coloured. b, Distance from filled gaps to the nearest segmental duplication (x axis) versus the counts of highly repetitive (>95%, green) and less repetitive (≤95%, orange) filled gaps in 100 base-pair bins (y axis). An additional 2,600 and 1,755 filled gaps map directly within segmental duplication sites with ≤95% and >95% repeat content, respectively. c, Polymorphism rates for lineage-specific MEIs. Alu, SVA, L1Pt and PTERV1 insertions that do not ‘lift over’ between chimpanzee and bonobo reference genomes were identified and genotyped for deletions using data from 10 bonobos and 10 chimpanzees. Light-coloured bars and percentages represent the fraction of instances of the MEI type that display support for polymorphism; dark-coloured bars represent the fraction of fixed insertions in these populations. PTERV1 displays a significantly less polymorphic fraction than Alu (P = 2.6 × 10−74, chimpanzee; P = 6.9 × 10−35, bonobo; χ2 test, Bonferroni correction), SVA (P = 3.8 × 10−19; P = 1.9 × 10−62) or L1Pt (P = 2.2 × 10−18; P = 1.3 × 10−8), reflecting its lack of activity since the divergence of Pan. SVA is the only MEI type with a greater polymorphism rate in bonobo. d, A COSEG network of bonobo-specific Alu subfamilies indicating the relative number of elements (size of the node) and number of mutations (line thickness) that distinguish subfamilies. e, A comparison of the retrotransposition rate per million years based on lineage-specific Alu insertions from a select panel of primate genomes. f–h, The percentage identity distribution (f) and length distribution (g) of segmental duplications (≥90% identify, ≥1 kb and no unplaced contigs) are shown as well as the pattern of the largest and most identical (≥10 kb and ≥98%) intrachromosomal (blue) and interchromosomal (red) segmental duplications (h) in the bonobo genome.