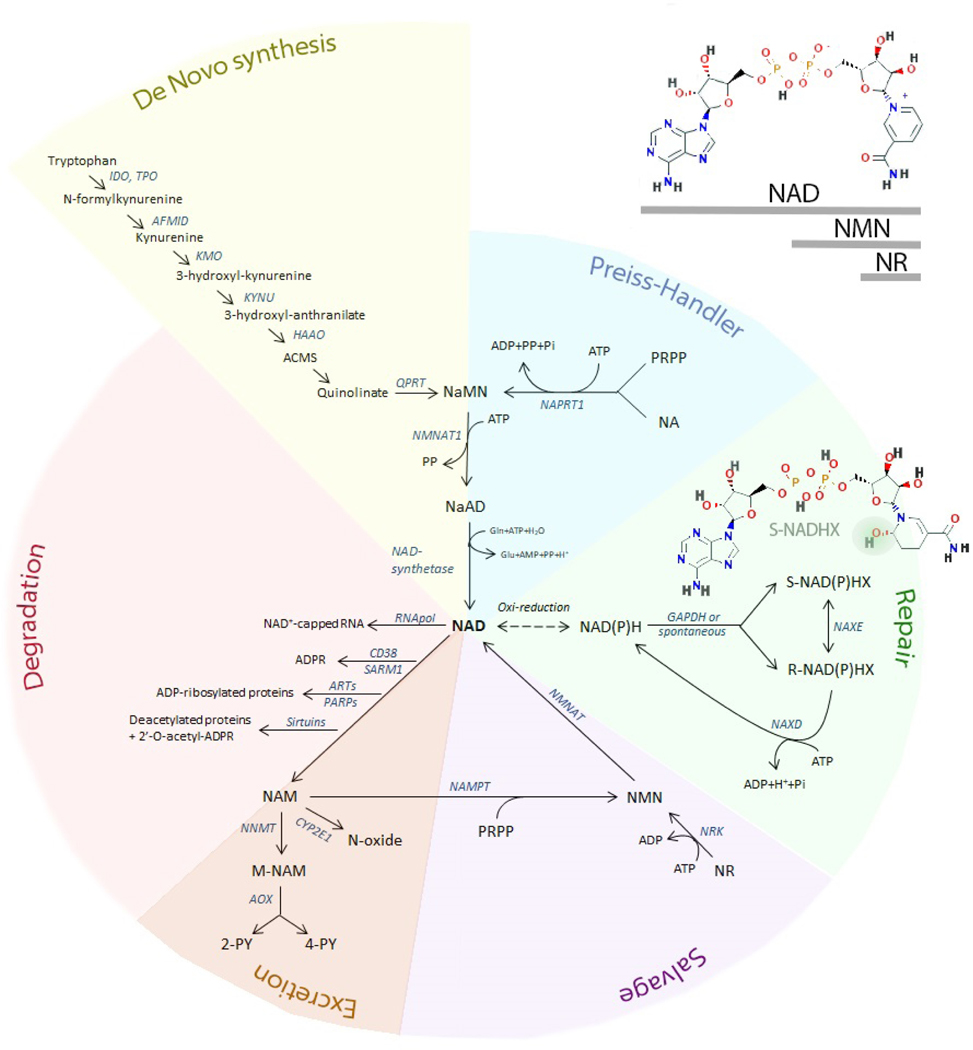
Figure 1. An overview of the NAD metabolome and metabolic pathways.
This figure is an integrative view of the networks related to NAD metabolism, including synthesis (de novo, Preiss-Handler, and salvage pathways), degradation, excretion, and repair pathways. The structure of NAD, NMN, and NR is shown (upper right). The structure of S-NADHX is shown (repair pathway - green) and the hydroxy group located at position 6 of the nicotinyl ring is highlighted. In the R-configuration the hydroxy group of the nicotinyl ring is located at position 2 (not shown). Cyclic forms of both S- and R-NAD(P)HX (not shown) are also toxic metabolites. 2-PY= N-methyl-2-pyridone-5-carboxamide; 4PY= N-methyl-4-pyridone-3-carboxamide; ACMS= 2-Amino-3-carboxymuconic acid semialdehyde; AFMID= Arylformamidase; AOX= aldehyde oxidase; ARTs= ADP-ribosyl-transferases; CYP2E1= Cytochrome P450 2E1; GAPDH= Glyceraldehyde-3-phosphate dehydrogenase; HAAO= 3-hydroxyanthranilate 3,4-dioxygenase; IDO= indoleamine 2,3-dioxygenase; KMO= kynurenine 3-monooxygenase; KYNU= kynureninase; M-NAM= methyl nicotinamide; NA= nicotinic acid; NaAD= nicotinic acid adenine dinucleotide; NAD= nicotinamide adenine dinucleotide; NAM= nicotinamide; NaMN= nicotinic acid mononucleotide; NAMPT= nicotinamide phosphoribosyltransferase; NaPRT1= nicotinic acid phosphoribosyltransferase 1; NAXD= NAD(P)HX Dehydratase; NAXE= NAD(P)H-hydrate epimerase; NMN= nicotinamide mononucleotide; NMNAT1= nicotinamide/nicotinic acid mononucleotide adenylyltransferase 1; NNMT= nicotinamide N-methyltranferase; NR= nicotinamide riboside; PARP= poly (ADP-ribose) polymerase; PRPP= phosphoribosyl pyrophosphate; QPRT= quinolinate phosphoribosyl transferase; SARM1= short for sterile alpha and Toll/interleukin receptor (TIR) motif-containing protein 1; TPO= tryptophan 2,3-dioxygenase;