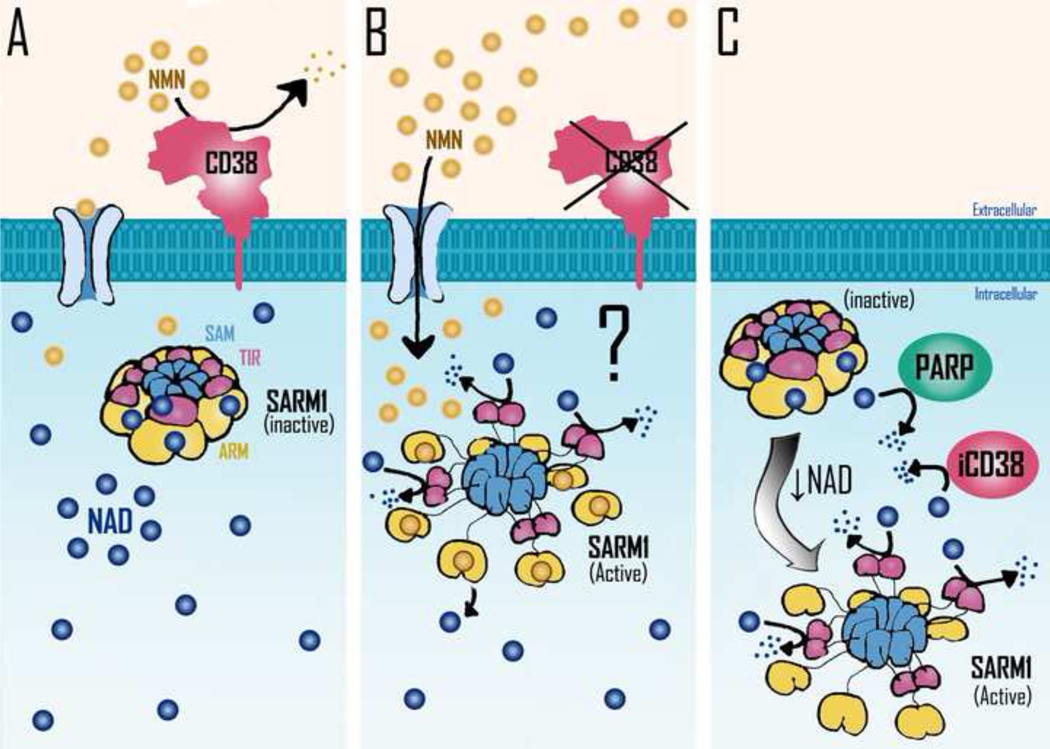
Figure 2. Potential mechanisms involved in Sarm1 activation.
SARM1 homo-octamer assumes a packed inactive conformation which is stabilized by NAD binding to allosteric sites located in the ARM domains. NAD decline leads to the disassembly of SARM1’s peripheral ARM ring, allowing the formation of TIR dimers, which are responsible for SARM1 NADase activity. It has been postulated that NMN may promote NAD displacement from SARM1 inhibitory allosteric sites, resulting in SARM1 NADase activation. A) In normal physiological context NAD is bound to allosteric sites in SARM1 oligomers, far from its catalytic sites. CD38 present in the cellular plasma membrane would lead to the degradation of extracellular NMN, preventing the increase of intracellular NMN levels. B) In a condition where extracellular CD38 activity is blocked, the consequent increase in intracellular NMN levels could lead to the displacement of NAD from SARM1’s allosteric inhibitory sites leading to SARM1 activation. C) Increased expression/activity of intracellular NADases such as PARPs or intracellular CD38 (iCD38) can lead to a decrease in intracellular NAD levels and consequent activation of SARM1 activity. Further decrease in NAD levels could lead to metabolic collapse and cell death. SAM= sterile alpha motif; TIR= toll/interleukin-1 receptor (TIR) homology domain; ARM= armadillo repeat domain.