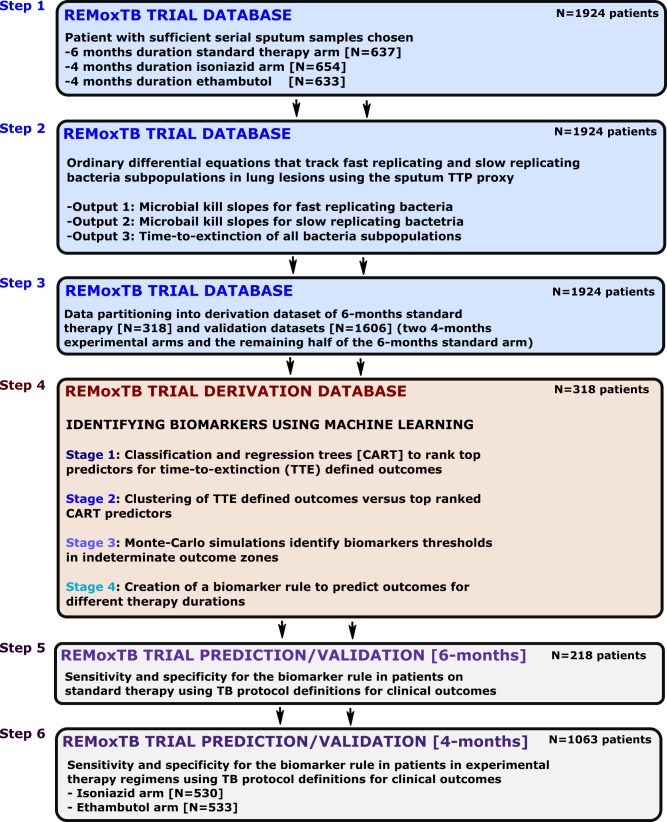
Fig. 1. Biomarker development steps.
Step 1: Patients without sufficient data points to derive bacterial kill slopes were removed. Step 2: The weekly sputum time-to-positivity data was then converted to colony forming units and then modeled using ordinary differential equations. Step 3: Data partitioning of 50% of patients in standard of care six-months therapy as derivation data-set and the other 50% into valdiation dataset. All patients in experimental arm, administered over 4 months were assigned to validation datasets. Step 4: Four mathematical modeling and machine learning types of analyses in derivation dataset to1 identify predictors of time-to-extinction (TTE) and2 threshold values deliniating different TTE, and3 design a diagnostic rule for different therapy durations. Step 5: Accuracy of diganostic rule/biomarker for six-months therapy duration in standard of care validation dataset using clinical definitions of outcome (relapse, cure). Step 6: Accuracy of diganostic rule/biomarker for four-months therapy duration in two experimental arms in validation dataset using clinical definitions of relapse and cure.