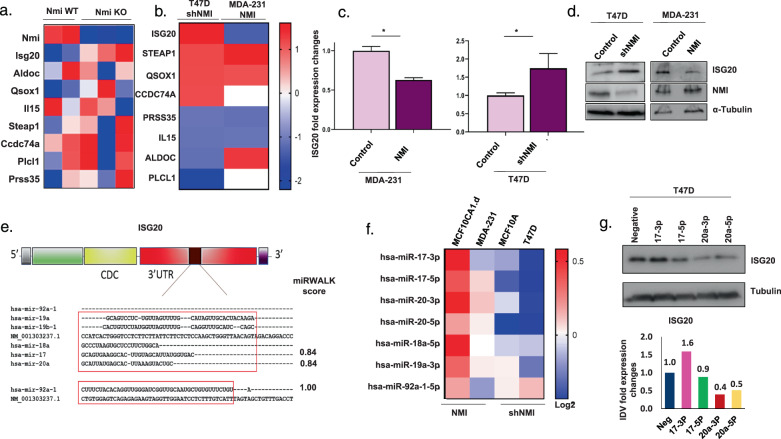
Fig. 5. NMI/STAT5A axis downregulate ISG20 through miRNA hsa-miR-20.
a Heatmap of RNA-seq expression of STAT5 enhancer related genes in mammary mouse tissues for wild-type (n = 2) vs NMI KO (n = 3) mice mammary tissue. b Heatmap of fold change RNA-seq expression of Stat5 enhancer related genes in T47D shNMI vs control and 231 NMI vs control. c Fold expression changes of ISG20 using RTq-PCR in T47D vector control and shNMI, MDA-MB-231 scrambled control and NMI, ISG20 expression was significantly higher in T47D shNMI (p = 0.03) and lower in MDA-MB-231 NMI (p ≤ 0.0001). d WB of ISG20 and NMI proteins in T47D control and shNMI and MDA-MB-231 control and NMI expressing cells. e Schematic illustration of ISG20 gene structure with predicted binding sites of miR-17–92 cluster at the 3’UTR region. f Heatmap representing fold changes of miR-17–92 cluster expression in miRNA array for MCF10CA1cl.D NMI, MDA-MB-231 NMI, MCF10A shNMI, and T47D shNMI compared to controls, scale represents log2 fold changes. g Western blot of ISG20 in T47D cells 48 h after being supplemented with miRNA mimics (100 nM) Hsa-miR-17-3p, Hsa-miR-17-5p, Hsa-miR-20a-3p, or Hsa-miR-20a-5p. Graph represents IDV quantification fold changes.